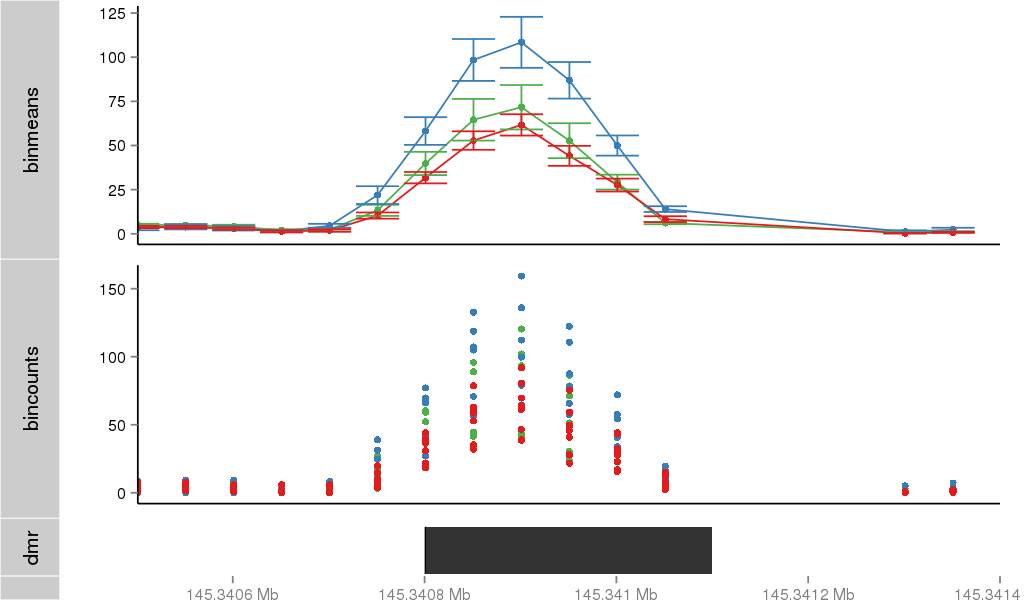
DMR 10018 chrX:145340801-145341100 (300bp)
| DMR Stats | |
|---|---|
| Position | chrX:145340801-145341100 (View on UCSC) |
| Width | 300bp |
| Genomic Context | intergenic |
| Nearest Genes | 5S_rRNA (167873bp away) |
| ANODEV p-value | 0.00462487525013 |
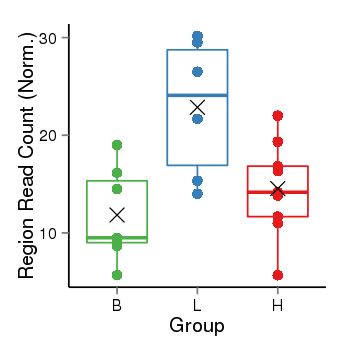
| Frequencies | Benign: 7.0 (100.0%), Low: 6.0 (100.0%), High: 9.0 (100.0 %) |
| Frequent? |
Region Count Boxplot
Region Overview
Gene Overlaps
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 10018 | chrX | 145340806 | 145340975 | wgEncodeRegDnaseClusteredV2 | 2 | score:227, srow:1278158 |
| 10018 | chrX | 145340664 | 145341259 | wgEncodeRegTfbsClusteredV3 | NFATC1 | score:414, expCount:1, expNums:96, expScores:414, srow:4371024 |
| 10018 | chrX | 145340781 | 145341084 | wgEncodeRegTfbsClusteredV3 | TCF3 | score:247, expCount:1, expNums:117, expScores:247, srow:4371025 |
| 10018 | chrX | 145340806 | 145341056 | wgEncodeRegTfbsClusteredV3 | BCL3 | score:485, expCount:1, expNums:83, expScores:485, srow:4371026 |
| 10018 | chrX | 145340818 | 145341037 | wgEncodeRegTfbsClusteredV3 | PBX3 | score:172, expCount:1, expNums:102, expScores:172, srow:4371027 |
| 10018 | chrX | 145340834 | 145341037 | wgEncodeRegTfbsClusteredV3 | MYC | score:237, expCount:1, expNums:625, expScores:237, srow:4371028 |
- 6 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 10018 | cellmeth_recounts | PrEC: 33, LNCaP: 43, DU145: 23 | 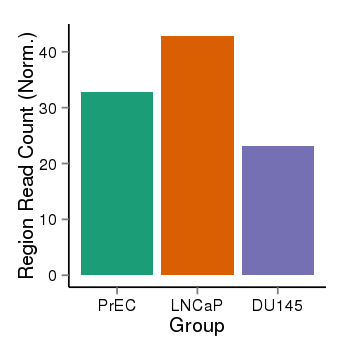 |
| 10018 | cellmeth_diffreps_PrECvsDU145 | A=49, B=16, Length=220, Event=Down, log2FC=-1.61, padj=0.00119638502767033 | 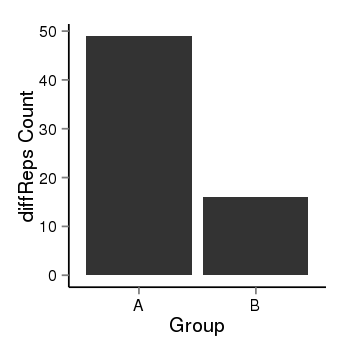 |
| 10018 | tcgaexp | gene=IL9R, entrez=3581, pos=chrX:59330252-155240482(+), B=6, L=4, M=6, H=9 | 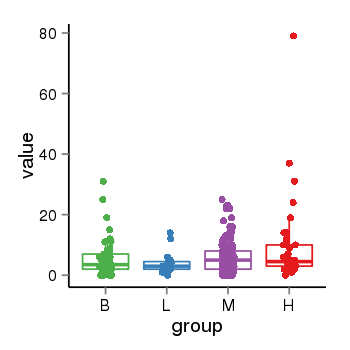 |
| 10018 | tcgaexp | gene=VAMP7, entrez=6845, pos=chrX:59213949-155173433(+), B=2541, L=2446, M=2565, H=2891 | 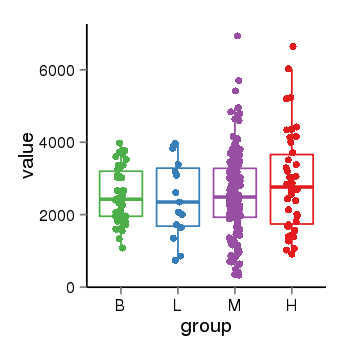 |
| 10018 | tcgaexp | gene=SPRY3, entrez=10251, pos=chrX:59100457-155012117(+), B=156, L=117, M=100, H=75 | 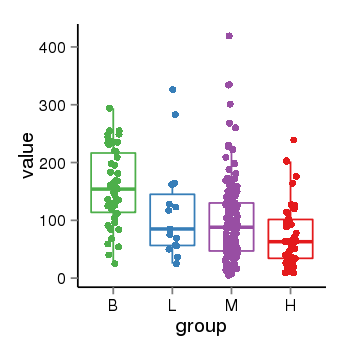 |