DMR 10287 chr1:15755351-15755500 (150bp)
| DMR Stats | |
|---|---|
| Position | chr1:15755351-15755500 (View on UCSC) |
| Width | 150bp |
| Genomic Context | exon |
| Nearest Genes | EFHD2 (0bp away) |
| ANODEV p-value | 0.0023829086855 |
| Frequencies | Benign: 5.0 (71.43%), Low: 1.0 (16.67%), High: 9.0 (100.0 %) |
| Frequent? |
Region Count Boxplot

Region Overview

Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10287 | EFHD2 | 271 | uc001awh.2 | chr1 | 15736391 | 15756839 | + | 150 | 100.0 | 0.82 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (100) | 0.0 |
- 1 genes
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 10287 | chr1 | 15755426 | 15755715 | wgEncodeRegDnaseClusteredV2 | 7 | score:225, srow:12038 |
| 10287 | chr1 | 15755363 | 15755365 | phastConsElements100way | lod=15 | score:262, srow:57160 |
| 10287 | chr1 | 15755416 | 15755418 | phastConsElements100way | lod=14 | score:255, srow:57161 |
- 3 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 10287 | cellmeth_recounts | PrEC: 20, LNCaP: 1, DU145: 3 |  |
| 10287 | cellmeth_diffreps_PrECvsLNCaP | A=142, B=7, Length=640, Event=Down, log2FC=-4.34, padj=0 |  |
| 10287 | cellmeth_diffreps_PrECvsDU145 | A=142, B=13, Length=600, Event=Down, log2FC=-3.45, padj=0 |  |
| 10287 | cellexp | id=ENSG00000142634, name=EFHD2, PrEC=14178, str=3492, LNCaP=1309, str=1182, DU145=35391, str=29693 |  |
| 10287 | tcgaexp | gene=SRSF10, entrez=10772, pos=chr1:36273-24306953(-), B=3487, L=3388, M=3432, H=4019 | 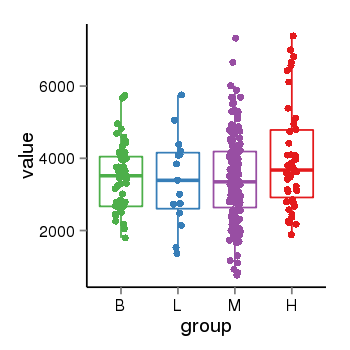 |
| 10287 | tcgaexp | gene=EFHD2, entrez=79180, pos=chr1:15736391-15756839(+), B=4654, L=4211, M=3493, H=2904 |  |
| 10287 | tcgaexp | gene=LOC100133331, entrez=100133331, pos=chr1:322037-180753553(-), B=268, L=328, M=271, H=305 | 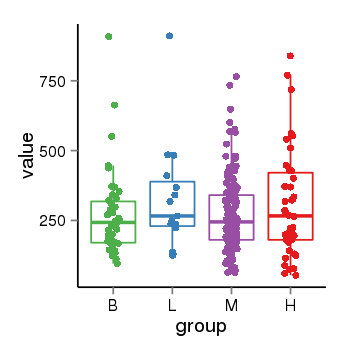 |