DMR 13433 chr4:8275901-8276050 (150bp)
| DMR Stats | |
|---|---|
| Position | chr4:8275901-8276050 (View on UCSC) |
| Width | 150bp |
| Genomic Context | intron |
| Nearest Genes | HTRA3 (0bp away) |
| ANODEV p-value | 0.0444476414348 |
| Frequencies | Benign: 6.0 (85.71%), Low: 2.0 (33.33%), High: 9.0 (100.0 %) |
| Frequent? |
Region Count Boxplot

Region Overview

Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13433 | HTRA3 | 21373 | uc003gkz.3 | chr4 | 8271489 | 8298200 | + | 150 | 100.0 | 0.24 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0) | 0.0 |
| 13433 | HTRA3 | 21373 | uc003gla.3 | chr4 | 8271489 | 8308838 | + | 150 | 100.0 | 0.24 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0), I8 (0), E9 (0) | 0.0 |
- 2 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 13433 | chr4 | 8275801 | 8276015 | wgEncodeRegDnaseClusteredV2 | 2 | score:116, srow:871666 |
| 13433 | chr4 | 8274092 | 8276091 | cpgShelf | — | values(x.gr)[overs$srow, ]:29759 |
- 2 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 13433 | cellmeth_recounts | PrEC: 19, LNCaP: 40, DU145: 0 |  |
| 13433 | cellmeth_diffreps_PrECvsDU145 | A=50, B=1, Length=660, Event=Down, log2FC=-5.64, padj=9.37725123848947e-13 |  |
| 13433 | cellmeth_diffreps_LNCaPvsDU145 | A=60.94, B=0.94, Length=680, Event=Down, log2FC=-6.03, padj=0 |  |
| 13433 | cellexp | id=ENSG00000170801, name=HTRA3, PrEC=3, str=7, LNCaP=8, str=7, DU145=110, str=44 |  |
| 13433 | tcgaexp | gene=UGT2B10, entrez=7365, pos=chr4:393964-69886113(+), B=12, L=1, M=2, H=8 | 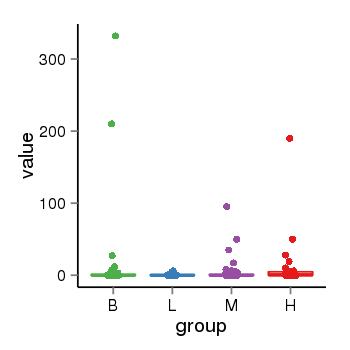 |
| 13433 | tcgaexp | gene=UGT2A3, entrez=79799, pos=chr4:506428-69817509(-), B=1, L=0, M=1, H=1 | 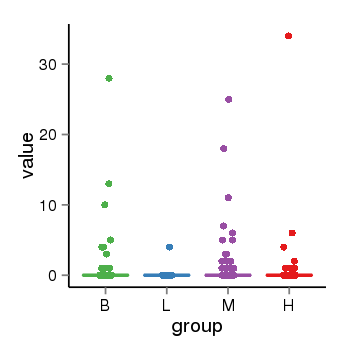 |
| 13433 | tcgaexp | gene=YTHDC1, entrez=91746, pos=chr4:6027-69215824(-), B=4512, L=3976, M=4109, H=4458 | 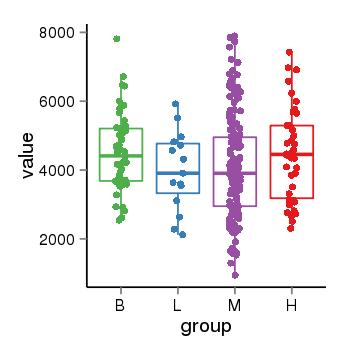 |
| 13433 | tcgaexp | gene=HTRA3, entrez=94031, pos=chr4:8271489-8308838(+), B=1063, L=1865, M=1186, H=1369 |  |
| 13433 | tcgaexp | gene=DUX4L5, entrez=653545, pos=chr4:72836-191010142(+), B=0, L=0, M=0, H=0 | 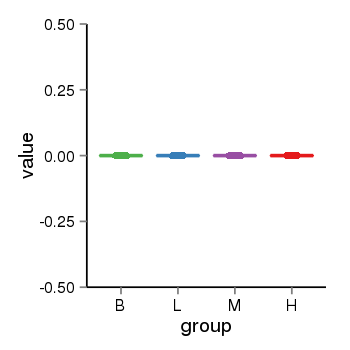 |