DMR 13495 chr4:30898701-30898900 (200bp)
| DMR Stats | |
|---|---|
| Position | chr4:30898701-30898900 (View on UCSC) |
| Width | 200bp |
| Genomic Context | intron |
| Nearest Genes | PCDH7 (0bp away) |
| ANODEV p-value | 0.000757166095324 |
| Frequencies | Benign: 5.0 (71.43%), Low: 0.0 (0.0%), High: 8.0 (88.89 %) |
| Frequent? |
Region Count Boxplot

Region Overview

Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13495 | PCDH7 | 21495 | uc011bxx.2 | chr4 | 30722037 | 31148423 | + | 200 | 100.0 | 5.36 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0) | 0.0 |
| 13495 | PCDH7 | 21495 | uc021xnd.1 | chr4 | 30722037 | 31148423 | + | 200 | 100.0 | 0.45 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0) | 0.0 |
- 2 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 13495 | chr4 | 30898721 | 30898955 | wgEncodeRegDnaseClusteredV2 | 2 | score:140, srow:879293 |
| 13495 | chr4 | 30898850 | 30898863 | tfbsConsSites | V$CEBPA_01 | score:878, zScore:1.71, srow:3737775 |
| 13495 | chr4 | 30898878 | 30898895 | tfbsConsSites | V$FOXJ2_01 | score:886, zScore:1.8, srow:3737776 |
| 13495 | chr4 | 30898881 | 30898895 | tfbsConsSites | V$HNF3B_01 | score:895, zScore:2.35, srow:3737777 |
- 4 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 13495 | cellmeth_recounts | PrEC: 1, LNCaP: 3, DU145: 1 |  |
| 13495 | cellexp | id=ENSG00000169851, name=PCDH7, PrEC=7422, str=9585, LNCaP=0, str=11, DU145=0, str=6 |  |
| 13495 | tcgaexp | gene=PCDH7, entrez=5099, pos=chr4:30722030-31148423(+), B=3961, L=1245, M=1476, H=1048 |  |
| 13495 | tcgaexp | gene=UGT2B10, entrez=7365, pos=chr4:393964-69886113(+), B=12, L=1, M=2, H=8 | 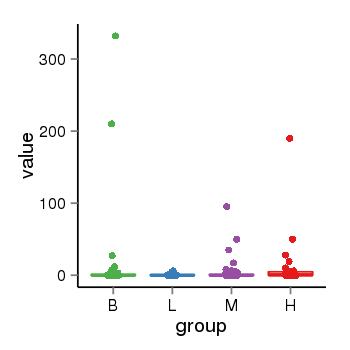 |
| 13495 | tcgaexp | gene=UGT2A3, entrez=79799, pos=chr4:506428-69817509(-), B=1, L=0, M=1, H=1 | 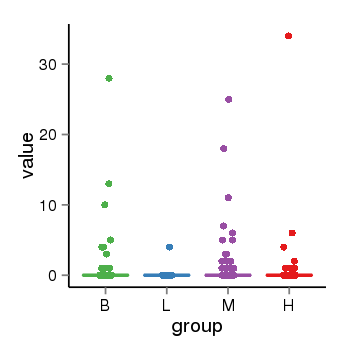 |
| 13495 | tcgaexp | gene=YTHDC1, entrez=91746, pos=chr4:6027-69215824(-), B=4512, L=3976, M=4109, H=4458 | 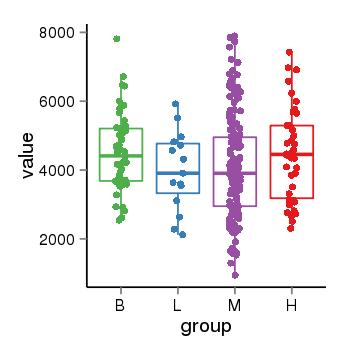 |
| 13495 | tcgaexp | gene=DUX4L5, entrez=653545, pos=chr4:72836-191010142(+), B=0, L=0, M=0, H=0 |  |