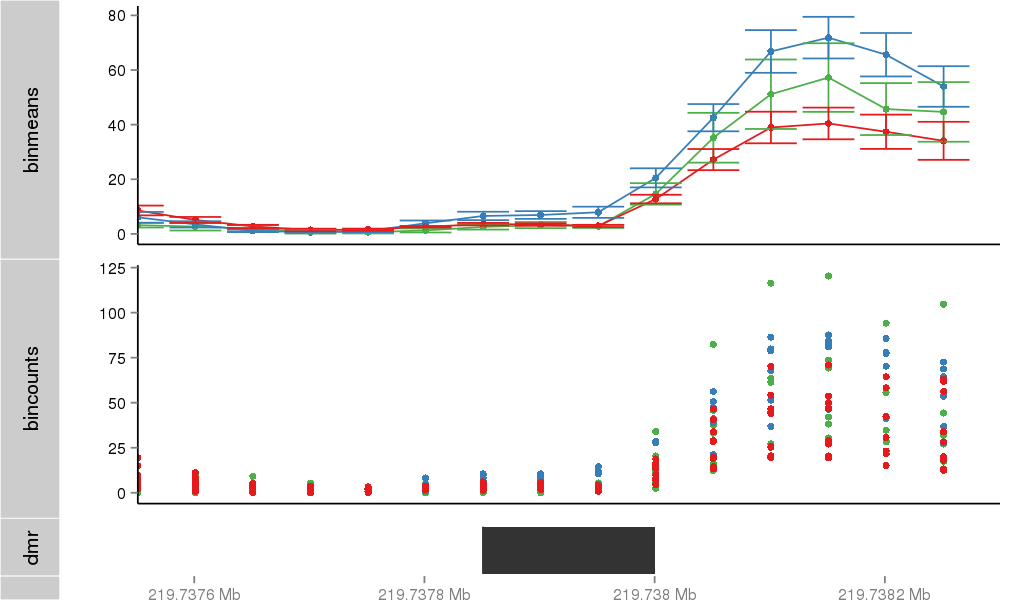
DMR 1385 chr2:219737851-219738000 (150bp)
| DMR Stats | |
|---|---|
| Position | chr2:219737851-219738000 (View on UCSC) |
| Width | 150bp |
| Genomic Context | 3' end |
| Nearest Genes | WNT6 (0bp away) |
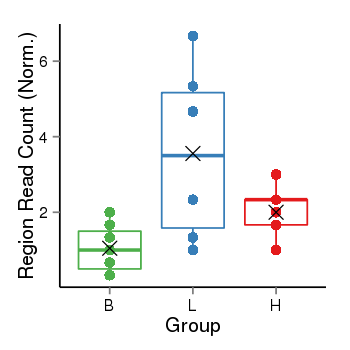
| ANODEV p-value | 0.0129322523589 |
| Frequencies | Benign: 0.0 (0.0%), Low: 3.0 (50.0%), High: 0.0 (0.0 %) |
| Frequent? |
Region Count Boxplot
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1385 | WNT6 | 17607 | uc002vjc.1 | chr2 | 219724546 | 219738954 | + | 150 | 100.0 | 0.63 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (100), E4 (0) | 30.67 |
- 1 genes
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 1385 | chr2 | 219737426 | 219738730 | wgEncodeRegDnaseClusteredV2 | 41 | score:1000, srow:683367 |
| 1385 | chr2 | 219737311 | 219739993 | wgEncodeRegTfbsClusteredV3 | EZH2 | score:1000, expCount:7, expNums:3,6,13,19,21,36,43, expScores:140,1000,343,99,439,385,536, srow:2393816 |
| 1385 | chr2 | 219737648 | 219737957 | wgEncodeRegTfbsClusteredV3 | SUZ12 | score:126, expCount:1, expNums:358, expScores:126, srow:2393819 |
| 1385 | chr2 | 219736593 | 219738081 | cpgShore | — | values(x.gr)[overs$srow, ]:28744 |
- 4 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 1385 | cellmeth_recounts | PrEC: 10, LNCaP: 1, DU145: 1 | 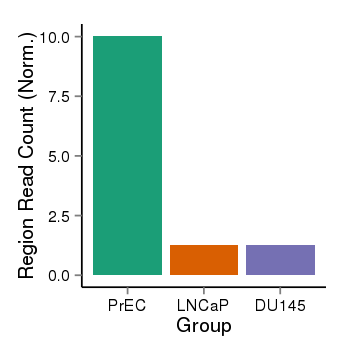 |
| 1385 | cellmeth_diffreps_PrECvsLNCaP | A=1266, B=102, Length=1100, Event=Down, log2FC=-3.63, padj=0 | 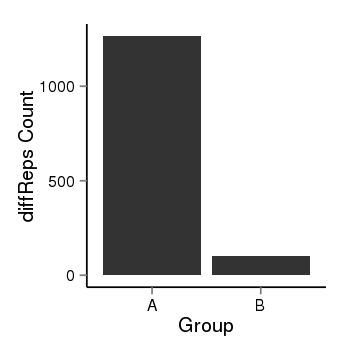 |
| 1385 | cellmeth_diffreps_PrECvsDU145 | A=1266, B=12, Length=1080, Event=Down, log2FC=-6.72, padj=0 | 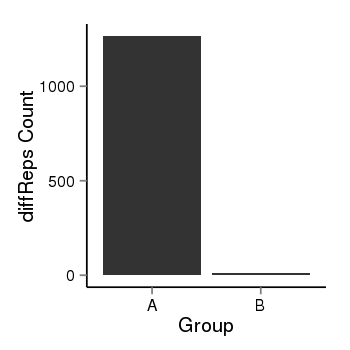 |
| 1385 | cellmeth_diffreps_LNCaPvsDU145 | A=109.04, B=10.29, Length=700, Event=Down, log2FC=-3.41, padj=0 | 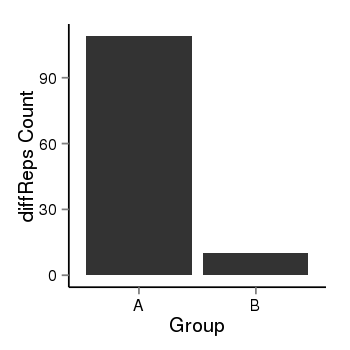 |
| 1385 | cellexp | id=ENSG00000115596, name=WNT6, PrEC=2, str=0, LNCaP=0, str=0, DU145=2, str=0 | 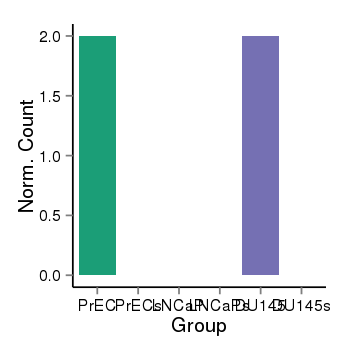 |
| 1385 | tcgaexp | gene=WNT6, entrez=7475, pos=chr2:219724546-219738954(+), B=87, L=66, M=48, H=31 | 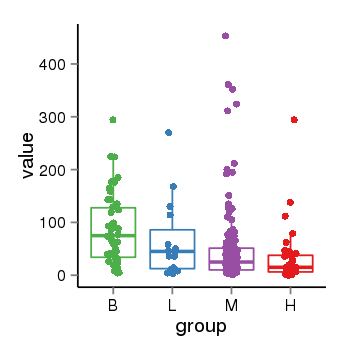 |