DMR 16219 chr8:10623501-10623650 (150bp)
| DMR Stats | |
|---|---|
| Position | chr8:10623501-10623650 (View on UCSC) |
| Width | 150bp |
| Genomic Context | 3' end |
| Nearest Genes | PINX1, SOX7 (0bp away) |
| ANODEV p-value | 0.0332022085687 |
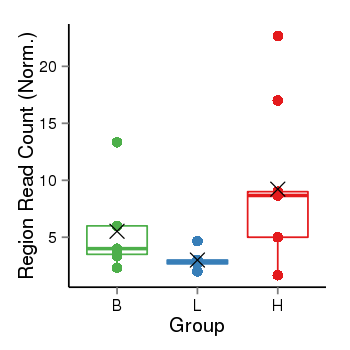
| Frequencies | Benign: 6.0 (85.71%), Low: 1.0 (16.67%), High: 8.0 (88.89 %) |
| Frequent? |
Region Count Boxplot
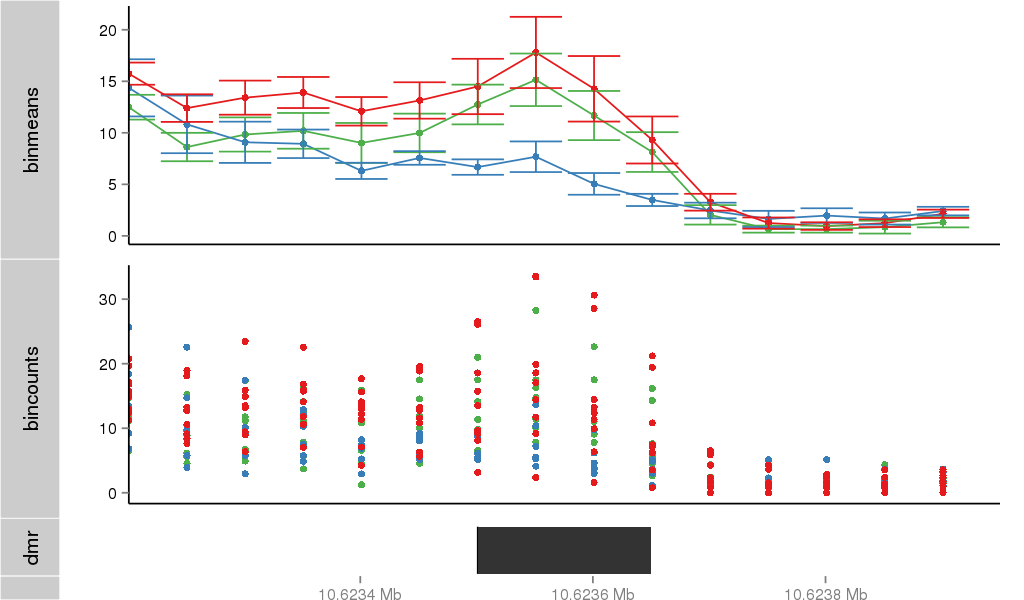
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16219 | PINX1 | 28169 | uc003wth.2 | chr8 | 10622884 | 10697299 | - | 150 | 100.0 | 0.76 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (100), E7 (0) | 100.0 |
| 16219 | PINX1 | 28169 | uc003wti.2 | chr8 | 10622884 | 10697299 | - | 150 | 100.0 | 0.76 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (100), E6 (0) | 100.0 |
| 16219 | SOX7 | 28169 | uc011kwz.2 | chr8 | 10581278 | 10697299 | - | 150 | 100.0 | 0.76 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (100), E6 (0) | 0.0 |
- 3 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 16219 | chr8 | 10623286 | 10623835 | wgEncodeRegDnaseClusteredV2 | 9 | score:225, srow:1141906 |
- 1 features
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 16219 | cellmeth_recounts | PrEC: 5, LNCaP: 8, DU145: 4 | 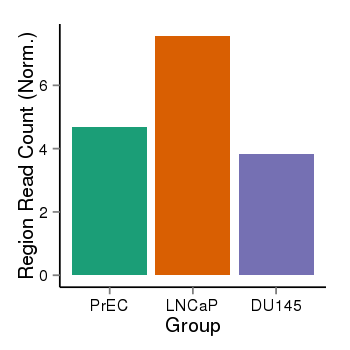 |
| 16219 | cellexp | id=ENSG00000171056, name=SOX7, PrEC=1817, str=995, LNCaP=10, str=1, DU145=34, str=28 | 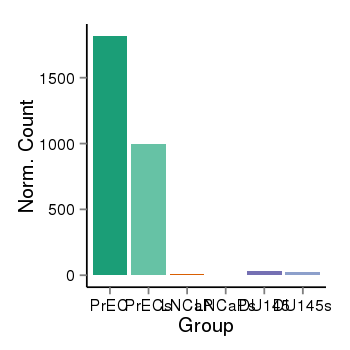 |
| 16219 | cellexp | id=ENSG00000248896, name=CTD-2135J3.3, PrEC=16, str=9, LNCaP=5, str=5, DU145=4, str=4 | 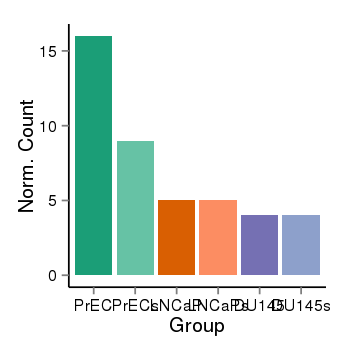 |
| 16219 | cellexp | id=ENSG00000254093, name=PINX1, PrEC=957, str=471, LNCaP=435, str=565, DU145=389, str=513 | 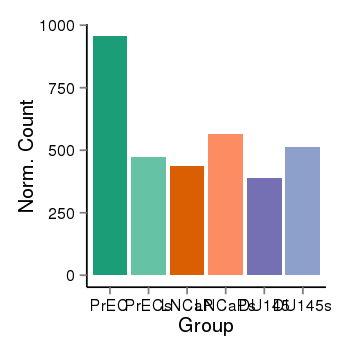 |
| 16219 | cellexp | id=ENSG00000258724, name=SOX7, PrEC=0, str=0, LNCaP=0, str=0, DU145=0, str=0 |  |
| 16219 | tcgaexp | gene=PINX1, entrez=54984, pos=chr8:10622884-10697299(-), B=227, L=229, M=233, H=217 | 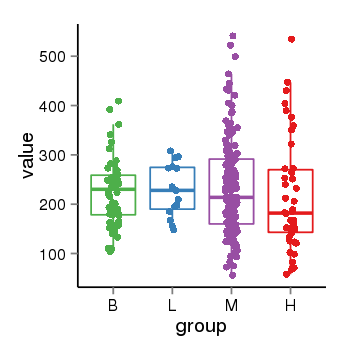 |
| 16219 | tcgaexp | gene=SOX7, entrez=83595, pos=chr8:10581278-10697299(-), B=1001, L=459, M=433, H=365 | 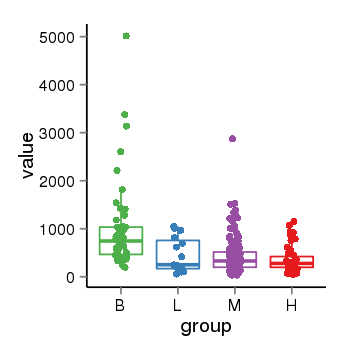 |