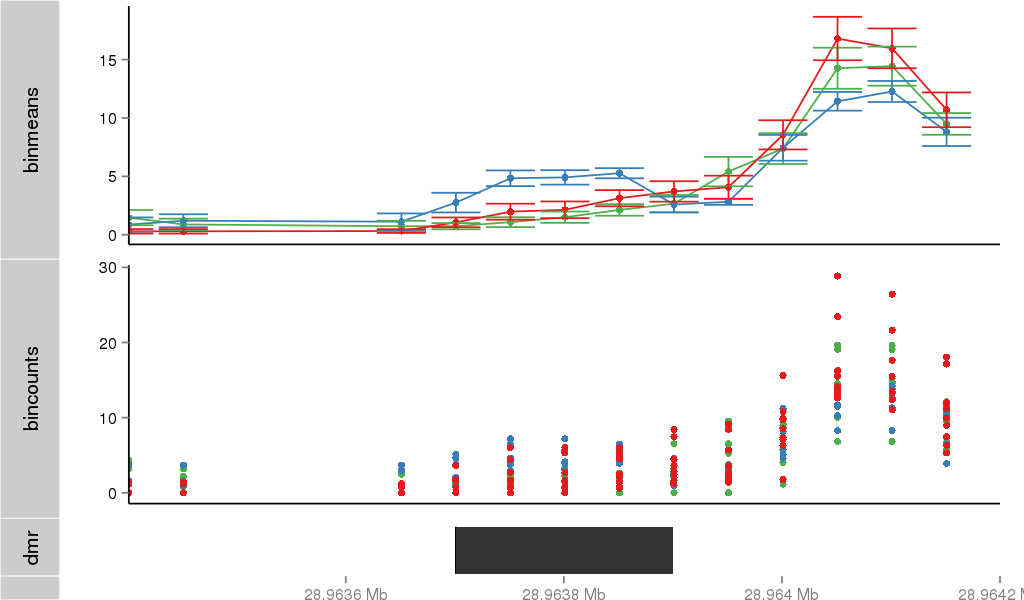
DMR 179 chr1:28963701-28963900 (200bp)
| DMR Stats | |
|---|---|
| Position | chr1:28963701-28963900 (View on UCSC) |
| Width | 200bp |
| Genomic Context | intron |
| Nearest Genes | TAF12 (0bp away) |
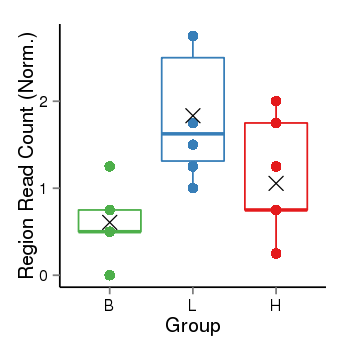
| ANODEV p-value | 0.0214886851926 |
| Frequencies | Benign: 0.0 (0.0%), Low: 0.0 (0.0%), High: 0.0 (0.0 %) |
| Frequent? |
Region Count Boxplot
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 179 | TAF12 | 541 | uc001bqx.3 | chr1 | 28929609 | 28969604 | - | 200 | 100.0 | 0.05 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0) | 0.0 |
| 179 | TAF12 | 541 | uc001bqy.3 | chr1 | 28929609 | 28969604 | - | 200 | 100.0 | 0.04 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0) | 0.0 |
| 179 | TAF12 | 541 | uc009vti.2 | chr1 | 28931451 | 28969604 | - | 200 | 100.0 | 0.07 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0) | 0.0 |
- 3 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 179 | chr1 | 28963606 | 28963795 | wgEncodeRegDnaseClusteredV2 | 2 | score:350, srow:23349 |
| 179 | chr1 | 28963860 | 28963869 | phastConsElements100way | lod=12 | score:240, srow:119952 |
- 2 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 179 | cellmeth_recounts | PrEC: 0, LNCaP: 3, DU145: 3 | 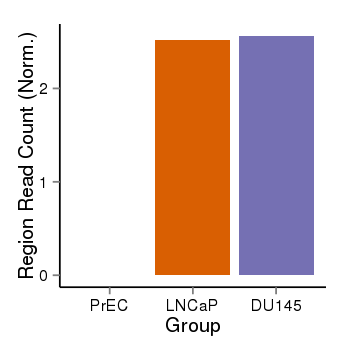 |
| 179 | cellexp | id=ENSG00000120656, name=TAF12, PrEC=1824, str=1267, LNCaP=1473, str=1484, DU145=1117, str=1053 | 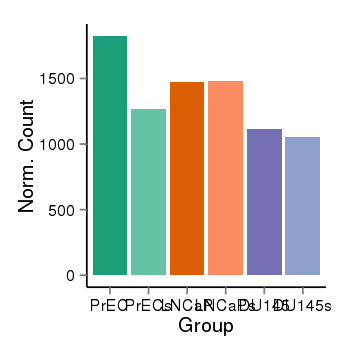 |
| 179 | tcgaexp | gene=TAF12, entrez=6883, pos=chr1:28929609-28969604(-), B=823, L=761, M=827, H=849 | 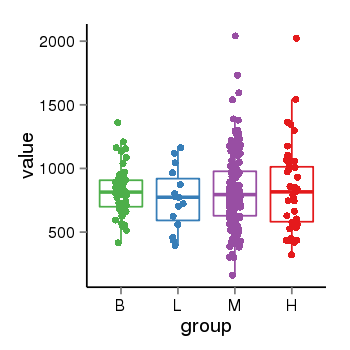 |
| 179 | tcgaexp | gene=LOC100133331, entrez=100133331, pos=chr1:322037-180753553(-), B=268, L=328, M=271, H=305 | 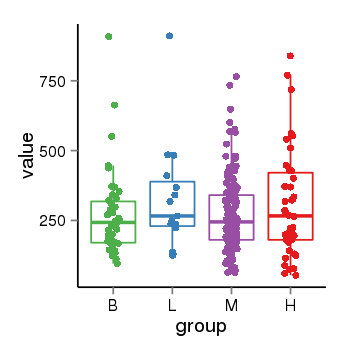 |