DMR 19562 chr12:124600101-124600300 (200bp)
| DMR Stats | |
|---|---|
| Position | chr12:124600101-124600300 (View on UCSC) |
| Width | 200bp |
| Genomic Context | intron |
| Nearest Genes | ZNF664-FAM101A (0bp away) |
| ANODEV p-value | 0.00863580287612 |
| Frequencies | Benign: 6.0 (85.71%), Low: 0.0 (0.0%), High: 8.0 (88.89 %) |
| Frequent? |
Region Count Boxplot

Region Overview

Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 19562 | ZNF664-FAM101A | 7197 | uc021rfy.1 | chr12 | 124457762 | 124800570 | + | 200 | 100.0 | 0.16 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (100), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0) | 0.0 |
- 1 genes
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 19562 | chr12 | 124600106 | 124600275 | wgEncodeRegDnaseClusteredV2 | 2 | score:156, srow:304187 |
| 19562 | chr12 | 124600112 | 124600116 | phastConsElements100way | lod=14 | score:255, srow:2265682 |
- 2 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 19562 | cellmeth_recounts | PrEC: 6, LNCaP: 0, DU145: 1 |  |
| 19562 | cellmeth_diffreps_PrECvsLNCaP | A=22, B=0, Length=400, Event=Down, log2FC=-Inf, padj=2.04501321634913e-06 |  |
| 19562 | cellmeth_diffreps_PrECvsDU145 | A=22, B=0, Length=400, Event=Down, log2FC=-Inf, padj=2.6934033431747e-06 |  |
| 19562 | cellexp | id=ENSG00000178882, name=FAM101A, PrEC=1, str=355, LNCaP=1, str=0, DU145=4, str=0 |  |
| 19562 | tcgaexp | gene=ZNF26, entrez=7574, pos=chr12:93510-133589154(+), B=419, L=392, M=412, H=512 | 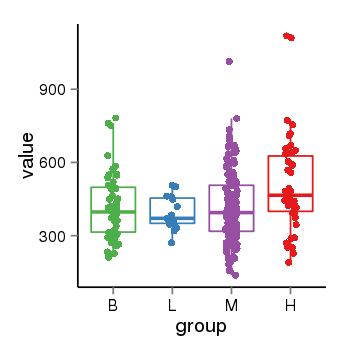 |
| 19562 | tcgaexp | gene=ZNF84, entrez=7637, pos=chr12:42780-133639885(+), B=1626, L=1649, M=1716, H=2045 | 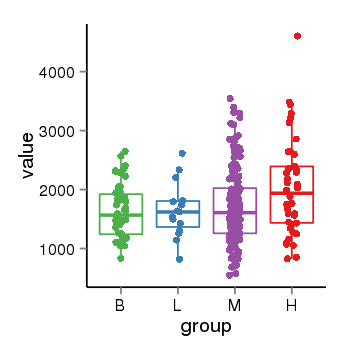 |