DMR 21580 chr17:6338451-6338600 (150bp)
| DMR Stats | |
|---|---|
| Position | chr17:6338451-6338600 (View on UCSC) |
| Width | 150bp |
| Genomic Context | promoter |
| Nearest Genes | AIPL1 (0bp away) |
| ANODEV p-value | 0.00275312606412 |
| Frequencies | Benign: 5.0 (71.43%), Low: 0.0 (0.0%), High: 8.0 (88.89 %) |
| Frequent? |
Region Count Boxplot

Region Overview

Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 21580 | AIPL1 | 12036 | uc002gcp.3 | chr17 | 6327059 | 6338519 | - | 69 | 46.0 | 0.08 | 0 | 100.0 | E1 (46), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0) | 0.0 |
| 21580 | AIPL1 | 12036 | uc002gcq.3 | chr17 | 6327059 | 6338519 | - | 69 | 46.0 | 0.08 | 0 | 100.0 | E1 (46), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0) | 0.0 |
| 21580 | AIPL1 | 12036 | uc002gcr.3 | chr17 | 6327059 | 6338519 | - | 69 | 46.0 | 0.08 | 0 | 100.0 | E1 (46), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0) | 0.0 |
| 21580 | AIPL1 | 12036 | uc002gcs.3 | chr17 | 6328641 | 6338519 | - | 69 | 46.0 | 0.08 | 0 | 100.0 | E1 (46), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0) | 0.0 |
| 21580 | AIPL1 | 12036 | uc010clk.3 | chr17 | 6327059 | 6338519 | - | 69 | 46.0 | 0.08 | 0 | 100.0 | E1 (46), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0) | 0.0 |
| 21580 | AIPL1 | 12036 | uc010cll.3 | chr17 | 6327059 | 6338519 | - | 69 | 46.0 | 0.08 | 0 | 100.0 | E1 (46), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0) | 0.0 |
| 21580 | AIPL1 | 12036 | uc021toq.1 | chr17 | 6327059 | 6338452 | - | 2 | 1.33 | 0.08 | 0 | 100.0 | E1 (1.33), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0) | 0.0 |
| 21580 | AIPL1 | 12036 | uc021tor.1 | chr17 | 6328641 | 6338519 | - | 69 | 46.0 | 0.08 | 0 | 100.0 | E1 (46), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0) | 0.0 |
- 8 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 21580 | chr17 | 6338386 | 6338615 | wgEncodeRegDnaseClusteredV2 | 6 | score:1000, srow:475865 |
| 21580 | chr17 | 6338462 | 6338468 | phastConsElements100way | lod=23 | score:304, srow:3550086 |
| 21580 | chr17 | 6338507 | 6338511 | phastConsElements100way | lod=23 | score:304, srow:3550087 |
- 3 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 21580 | cellmeth_recounts | PrEC: 3, LNCaP: 0, DU145: 0 |  |
| 21580 | cellexp | id=ENSG00000129221, name=AIPL1, PrEC=0, str=0, LNCaP=0, str=0, DU145=0, str=1 |  |
| 21580 | tcgameth | site:cg02482001, B=0.57, L=0.53, H=0.52 |  |
| 21580 | tcgaexp | gene=CRHR1, entrez=1394, pos=chr17:954315-43913194(+), B=36, L=21, M=25, H=18 | 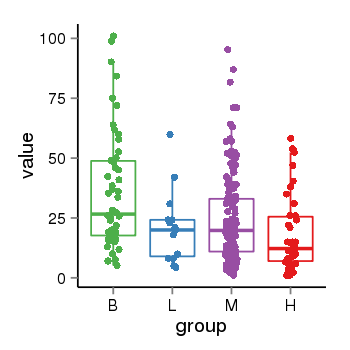 |
| 21580 | tcgaexp | gene=MAPT, entrez=4137, pos=chr17:762281-44105699(+), B=1933, L=2382, M=2487, H=2583 | 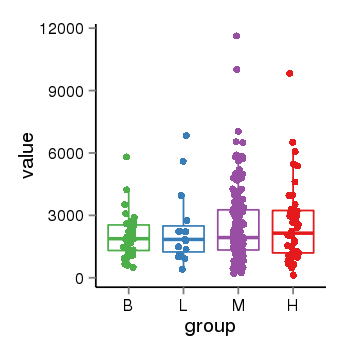 |
| 21580 | tcgaexp | gene=NSF, entrez=4905, pos=chr17:266212-44834828(+), B=3355, L=5678, M=5410, H=5700 | 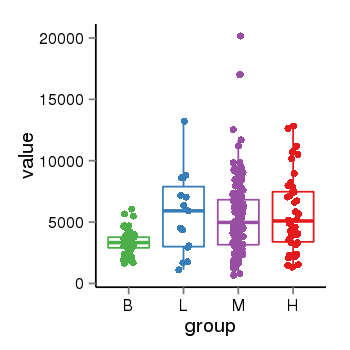 |
| 21580 | tcgaexp | gene=PLEKHM1, entrez=9842, pos=chr17:128328-43568146(-), B=1993, L=1631, M=1589, H=1418 | 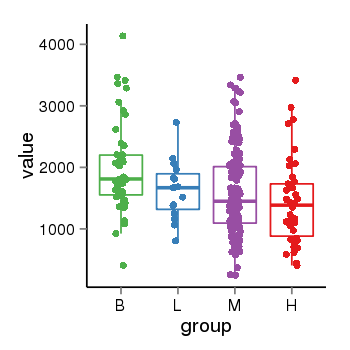 |
| 21580 | tcgaexp | gene=LRRC37A, entrez=9884, pos=chr17:415627-44415160(+), B=924, L=896, M=971, H=940 | 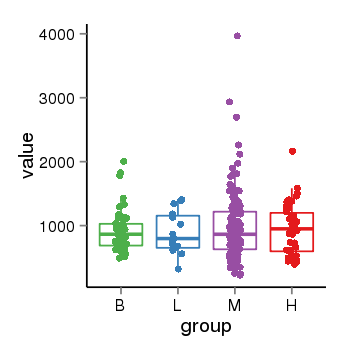 |
| 21580 | tcgaexp | gene=AIPL1, entrez=23746, pos=chr17:6327059-6338519(-), B=1, L=1, M=0, H=0 |  |
| 21580 | tcgaexp | gene=ARL17A, entrez=51326, pos=chr17:195721-44657088(-), B=200, L=195, M=210, H=230 | 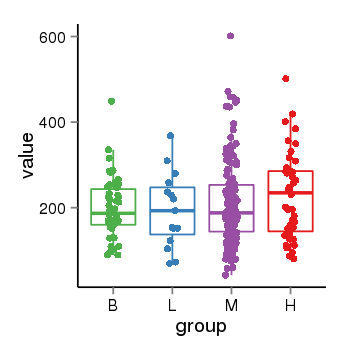 |
| 21580 | tcgaexp | gene=LRRC37A4P, entrez=55073, pos=chr17:197706-43597889(-), B=1015, L=1512, M=1396, H=1450 | 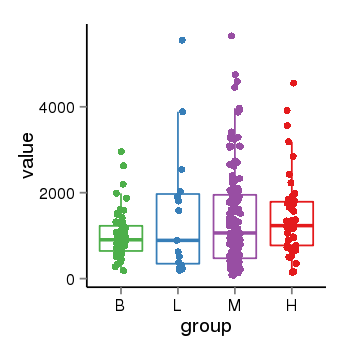 |
| 21580 | tcgaexp | gene=CRHR1-IT1, entrez=147081, pos=chr17:1143726-43723595(+), B=197, L=211, M=215, H=223 | 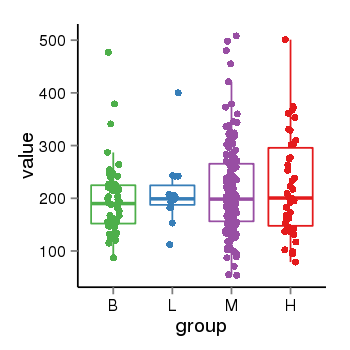 |
| 21580 | tcgaexp | gene=SPPL2C, entrez=162540, pos=chr17:943102-43924438(+), B=0, L=0, M=0, H=0 | 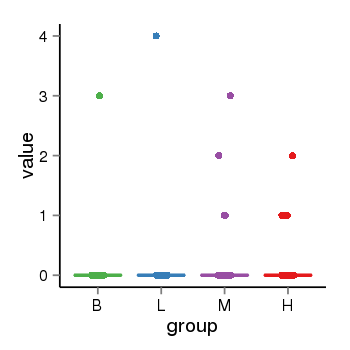 |
| 21580 | tcgaexp | gene=ARHGAP27, entrez=201176, pos=chr17:86404-43511112(-), B=1570, L=1264, M=1273, H=1486 | 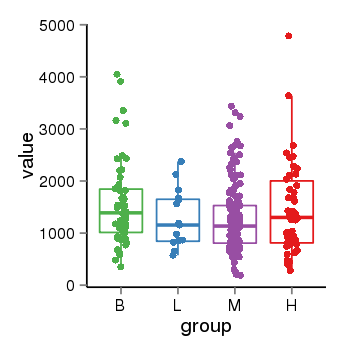 |
| 21580 | tcgaexp | gene=STH, entrez=246744, pos=chr17:790689-44077060(+), B=0, L=0, M=0, H=0 | 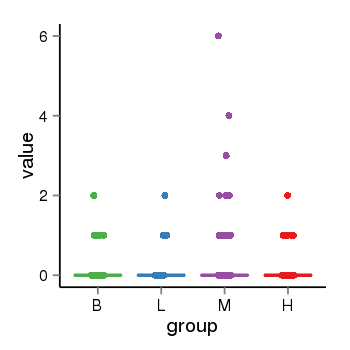 |
| 21580 | tcgaexp | gene=KANSL1, entrez=284058, pos=chr17:563498-44302740(-), B=3141, L=2594, M=2797, H=3017 | 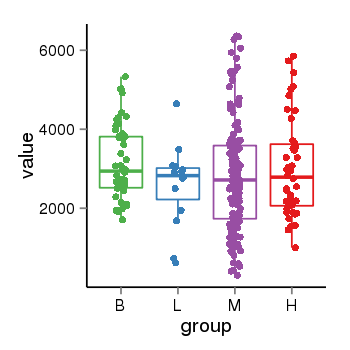 |
| 21580 | tcgaexp | gene=MGC57346, entrez=401884, pos=chr17:1151952-43715329(+), B=601, L=581, M=566, H=598 | 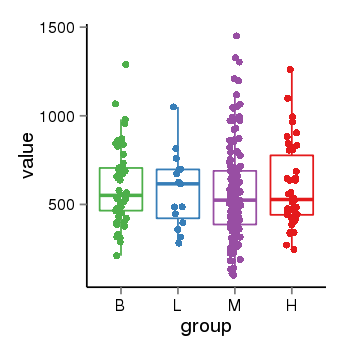 |
| 21580 | tcgaexp | gene=LRRC37A2, entrez=474170, pos=chr17:197706-44633014(+), B=351, L=310, M=361, H=377 | 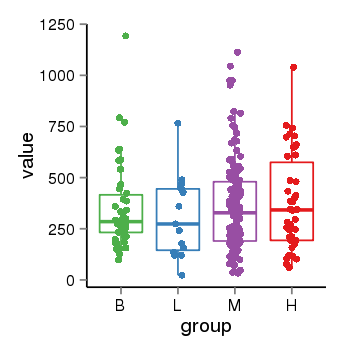 |
| 21580 | tcgaexp | gene=LOC644172, entrez=644172, pos=chr17:1188439-43679748(-), B=84, L=82, M=93, H=90 | 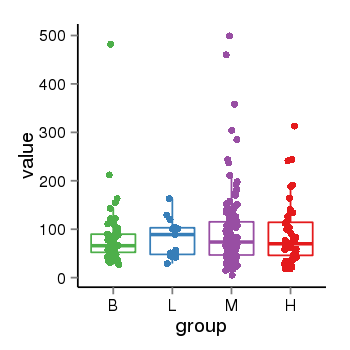 |
| 21580 | tcgaexp | gene=MAPT-AS1, entrez=100128977, pos=chr17:894694-43972879(-), B=1, L=2, M=4, H=2 | 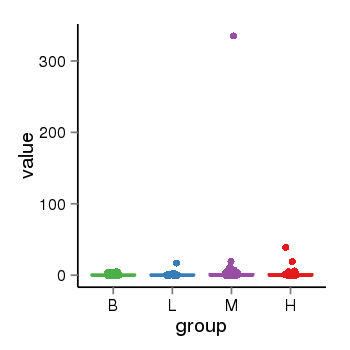 |
| 21580 | tcgaexp | gene=MAPT-IT1, entrez=100130148, pos=chr17:891434-43976164(+), B=5, L=3, M=3, H=4 | 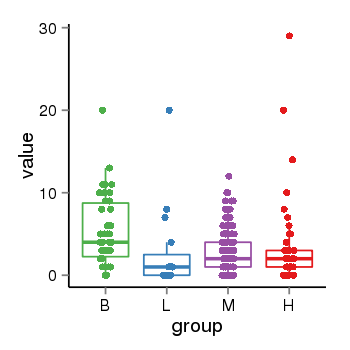 |