DMR 24864 chr1:906451-906600 (150bp)
| DMR Stats | |
|---|---|
| Position | chr1:906451-906600 (View on UCSC) |
| Width | 150bp |
| Genomic Context | exon |
| Nearest Genes | PLEKHN1 (0bp away) |
| ANODEV p-value | 0.00635159778583 |
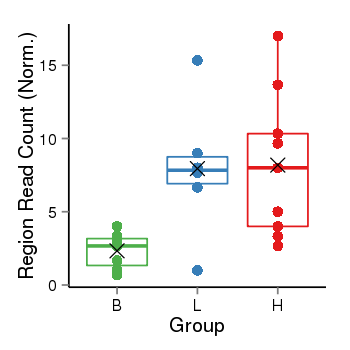
| Frequencies | Benign: 2.0 (28.57%), Low: 5.0 (83.33%), High: 8.0 (88.89 %) |
| Frequent? |
Region Count Boxplot
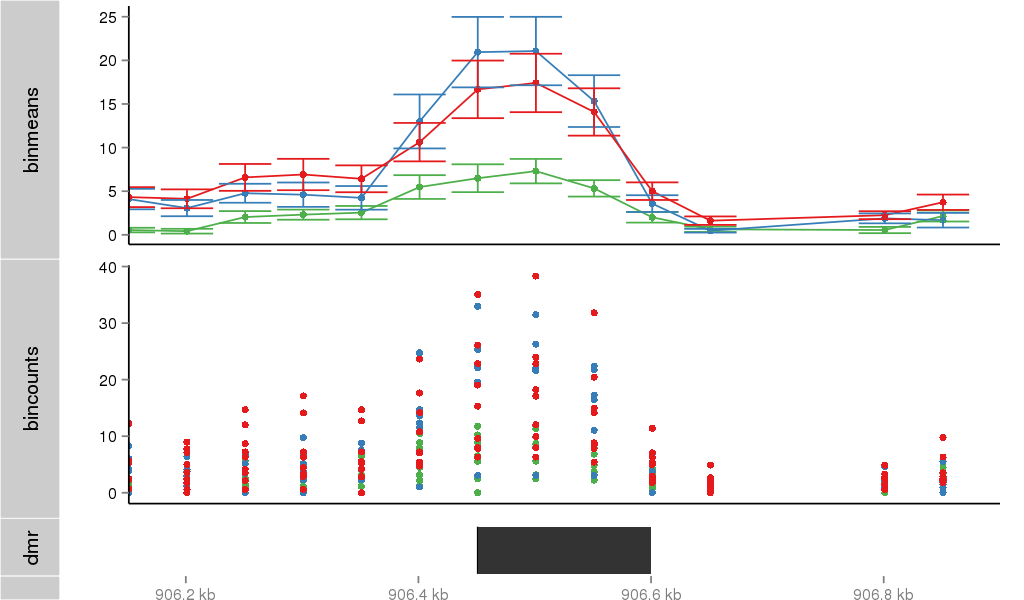
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 24864 | PLEKHN1 | 33 | uc001acd.3 | chr1 | 901877 | 910484 | + | 150 | 100.0 | 1.78 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (28), E7 (64), I7 (8.67), E8 (0), I8 (0), E9 (0), I9 (0), E10 (0), I10 (0), E11 (0), I11 (0), E12 (0), I12 (0), E13 (0), I13 (0), E14 (0), I14 (0), E15 (0), I15 (0), E16 (0) | 0.0 |
| 24864 | PLEKHN1 | 33 | uc001ace.3 | chr1 | 901877 | 910484 | + | 150 | 100.0 | 1.52 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (4), E6 (88), I6 (8.67), E7 (0), I7 (0), E8 (0), I8 (0), E9 (0), I9 (0), E10 (0), I10 (0), E11 (0), I11 (0), E12 (0), I12 (0), E13 (0), I13 (0), E14 (0), I14 (0), E15 (0) | 0.0 |
| 24864 | PLEKHN1 | 33 | uc001acf.3 | chr1 | 901877 | 910484 | + | 150 | 100.0 | 1.52 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (4), E7 (88), I7 (8.67), E8 (0), I8 (0), E9 (0), I9 (0), E10 (0), I10 (0), E11 (0), I11 (0), E12 (0), I12 (0), E13 (0), I13 (0), E14 (0), I14 (0), E15 (0) | 0.0 |
- 3 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 24864 | chr1 | 906366 | 906655 | wgEncodeRegDnaseClusteredV2 | 3 | score:310, srow:176 |
| 24864 | chr1 | 906538 | 906556 | tfbsConsSites | V$SEF1_C | score:730, zScore:1.87, srow:128 |
| 24864 | chr1 | 906539 | 906557 | tfbsConsSites | V$GR_Q6 | score:823, zScore:1.68, srow:129 |
| 24864 | chr1 | 906549 | 906569 | tfbsConsSites | V$PPARG_01 | score:728, zScore:1.71, srow:130 |
| 24864 | chr1 | 906566 | 906587 | tfbsConsSites | V$HEN1_02 | score:760, zScore:1.81, srow:131 |
| 24864 | chr1 | 906571 | 906582 | tfbsConsSites | V$LMO2COM_01 | score:923, zScore:1.76, srow:132 |
| 24864 | chr1 | 906572 | 906581 | tfbsConsSites | V$MYOD_Q6 | score:967, zScore:1.81, srow:133 |
| 24864 | chr1 | 906512 | 906512 | cosmic | COSM1185262 | srow:118 |
| 24864 | chr1 | 906515 | 906516 | cosmic | COSM1602769 | srow:119 |
| 24864 | chr1 | 906515 | 906516 | cosmic | COSM1602768 | srow:120 |
| 24864 | chr1 | 906518 | 906518 | cosmic | COSM913106 | srow:121 |
| 24864 | chr1 | 906532 | 906532 | cosmic | COSM1221180 | srow:122 |
| 24864 | chr1 | 906536 | 906536 | cosmic | COSM1344778 | srow:123 |
| 24864 | chr1 | 906455 | 906458 | phastConsElements100way | lod=18 | score:280, srow:2593 |
| 24864 | chr1 | 906472 | 906476 | phastConsElements100way | lod=25 | score:312, srow:2594 |
| 24864 | chr1 | 906478 | 906483 | phastConsElements100way | lod=21 | score:295, srow:2595 |
| 24864 | chr1 | 906538 | 906539 | phastConsElements100way | lod=20 | score:290, srow:2596 |
| 24864 | chr1 | 906548 | 906554 | phastConsElements100way | lod=23 | score:304, srow:2597 |
| 24864 | chr1 | 906560 | 906584 | phastConsElements100way | lod=105 | score:454, srow:2598 |
| 24864 | chr1 | 906586 | 906590 | phastConsElements100way | lod=33 | score:340, srow:2599 |
| 24864 | chr1 | 906297 | 906538 | cpgIsland | CpG: 23 | length:242, cpgNum:23, gcNum:175, perCpg:19, perGc:72.3, obsExp:0.74, srow:21 |
| 24864 | chr1 | 906539 | 908538 | cpgShore | — | values(x.gr)[overs$srow, ]:37 |
- 22 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 24864 | cellmeth_recounts | PrEC: 4, LNCaP: 34, DU145: 10 | 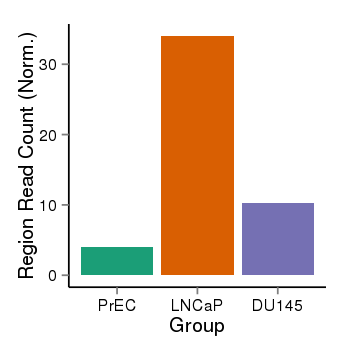 |
| 24864 | cellmeth_diffreps_PrECvsLNCaP | A=7, B=81, Length=840, Event=Up, log2FC=3.53, padj=0 | 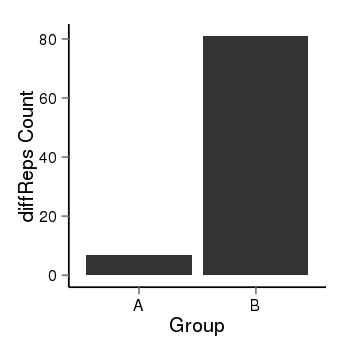 |
| 24864 | cellmeth_diffreps_LNCaPvsDU145 | A=99.42, B=7.48, Length=860, Event=Down, log2FC=-3.73, padj=0 | 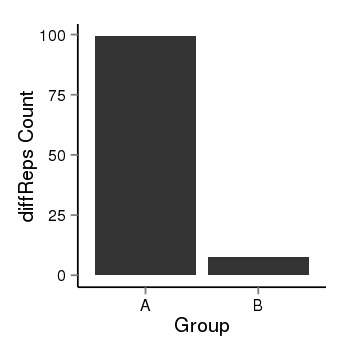 |
| 24864 | cellexp | id=ENSG00000187583, name=PLEKHN1, PrEC=1198, str=1239, LNCaP=961, str=487, DU145=492, str=700 | 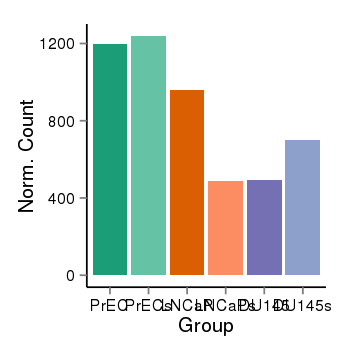 |
| 24864 | tcgameth | site:cg22481263, B=0.37, L=0.61, H=0.59 | 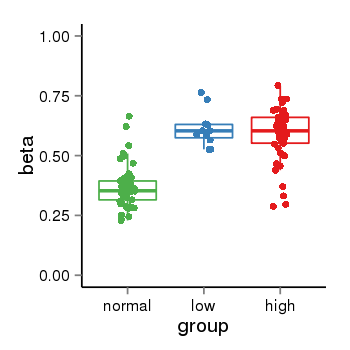 |
| 24864 | tcgaexp | gene=SRSF10, entrez=10772, pos=chr1:36273-24306953(-), B=3487, L=3388, M=3432, H=4019 | 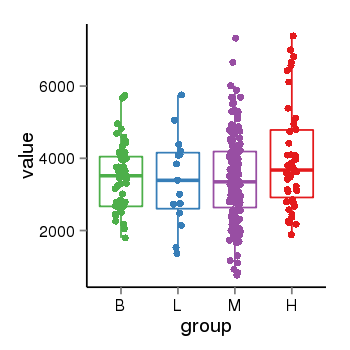 |
| 24864 | tcgaexp | gene=PLEKHN1, entrez=84069, pos=chr1:901877-910484(+), B=115, L=171, M=163, H=237 | 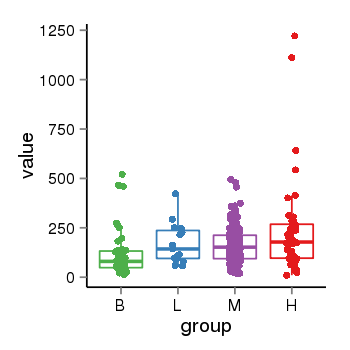 |
| 24864 | tcgaexp | gene=LOC100133331, entrez=100133331, pos=chr1:322037-180753553(-), B=268, L=328, M=271, H=305 | 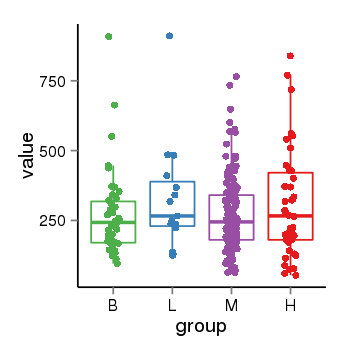 |