DMR 24902 chr1:2387451-2387600 (150bp)
| DMR Stats | |
|---|---|
| Position | chr1:2387451-2387600 (View on UCSC) |
| Width | 150bp |
| Genomic Context | intergenic |
| Nearest Genes | PLCH2 (20153bp away) |
| ANODEV p-value | 3.72077391906e-05 |
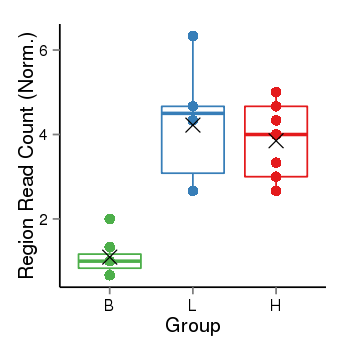
| Frequencies | Benign: 0.0 (0.0%), Low: 4.0 (66.67%), High: 6.0 (66.67 %) |
| Frequent? |
Region Count Boxplot
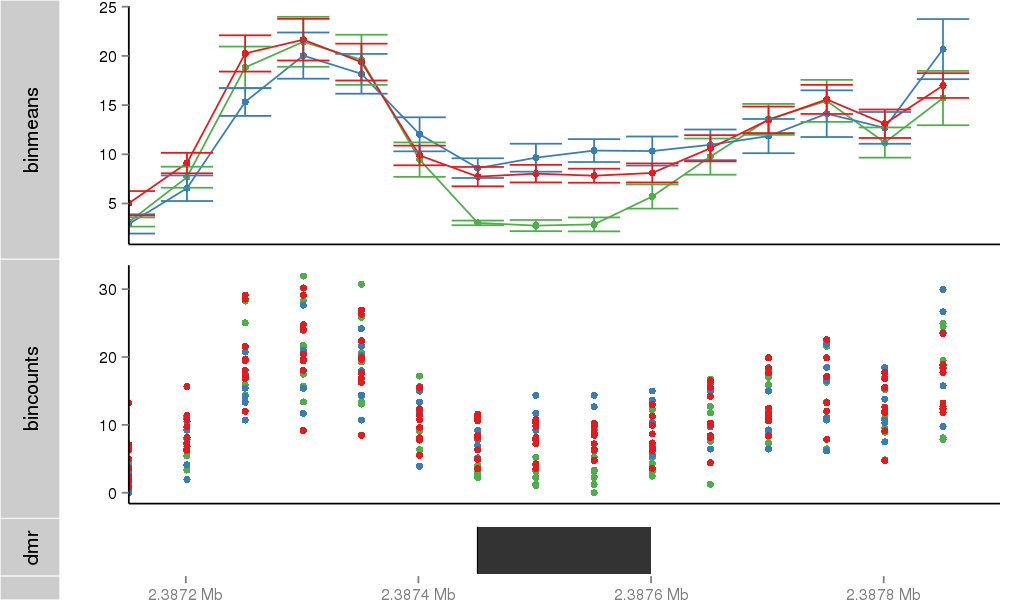
Region Overview
Gene Overlaps
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 24902 | chr1 | 2387321 | 2387550 | wgEncodeRegDnaseClusteredV2 | 14 | score:744, srow:1593 |
| 24902 | chr1 | 2387249 | 2387718 | wgEncodeRegTfbsClusteredV3 | POLR2A | score:217, expCount:1, expNums:249, expScores:217, srow:7549 |
| 24902 | chr1 | 2387353 | 2387722 | wgEncodeRegTfbsClusteredV3 | CTCF | score:190, expCount:3, expNums:18,597,635, expScores:124,190,159, srow:7550 |
| 24902 | chr1 | 2387457 | 2387489 | phastConsElements100way | lod=77 | score:423, srow:10561 |
| 24902 | chr1 | 2385670 | 2387669 | cpgShore | — | values(x.gr)[overs$srow, ]:317 |
- 5 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 24902 | cellmeth_recounts | PrEC: 4, LNCaP: 6, DU145: 4 | 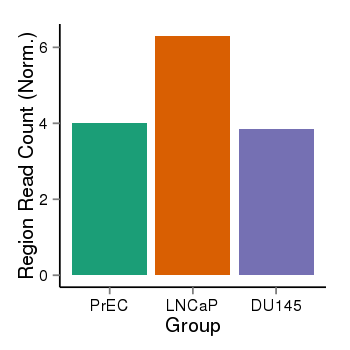 |
| 24902 | cellmeth_diffreps_PrECvsLNCaP | A=219, B=7, Length=720, Event=Down, log2FC=-4.97, padj=0 |  |
| 24902 | cellmeth_diffreps_PrECvsDU145 | A=219, B=11, Length=720, Event=Down, log2FC=-4.32, padj=0 | 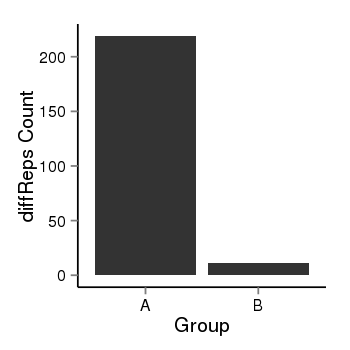 |
| 24902 | cellexp | id=ENSG00000149527, name=PLCH2, PrEC=15605, str=15786, LNCaP=478, str=150, DU145=59, str=40 | 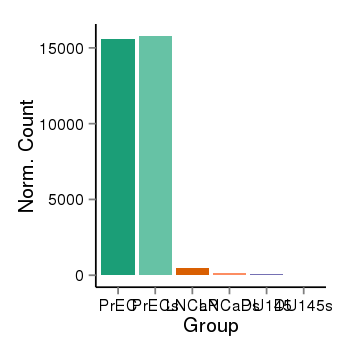 |
| 24902 | tcgaexp | gene=SRSF10, entrez=10772, pos=chr1:36273-24306953(-), B=3487, L=3388, M=3432, H=4019 | 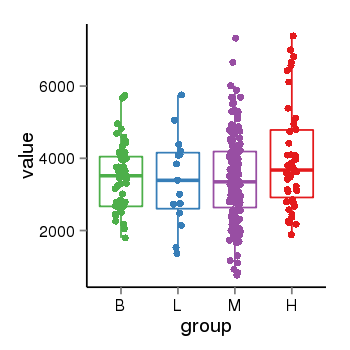 |
| 24902 | tcgaexp | gene=LOC100133331, entrez=100133331, pos=chr1:322037-180753553(-), B=268, L=328, M=271, H=305 | 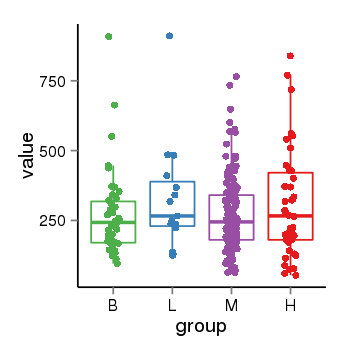 |