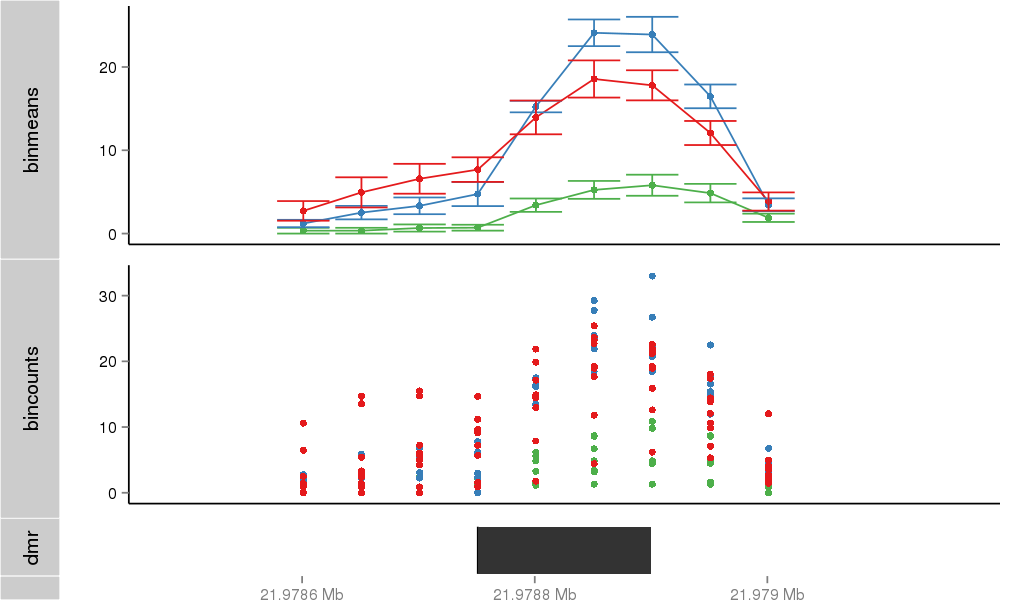
DMR 25121 chr1:21978751-21978900 (150bp)
| DMR Stats | |
|---|---|
| Position | chr1:21978751-21978900 (View on UCSC) |
| Width | 150bp |
| Genomic Context | promoter |
| Nearest Genes | RAP1GAP (0bp away) |
| ANODEV p-value | 1.6596797936e-09 |
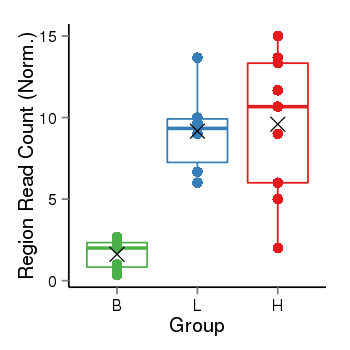
| Frequencies | Benign: 0.0 (0.0%), Low: 6.0 (100.0%), High: 8.0 (88.89 %) |
| Frequent? |
Region Count Boxplot
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 25121 | RAP1GAP | 394 | uc001bex.3 | chr1 | 21922708 | 21995856 | - | 150 | 100.0 | 0.13 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0), I8 (0), E9 (0), I9 (0), E10 (0), I10 (0), E11 (0), I11 (0), E12 (0), I12 (0), E13 (0), I13 (0), E14 (0), I14 (0), E15 (0), I15 (0), E16 (0), I16 (0), E17 (0), I17 (0), E18 (0), I18 (0), E19 (0), I19 (0), E20 (0), I20 (0), E21 (0), I21 (0), E22 (0), I22 (0), E23 (0), I23 (0), E24 (0), I24 (0), E25 (0) | 0.0 |
| 25121 | RAP1GAP | 394 | uc001bey.3 | chr1 | 21922708 | 21995856 | - | 150 | 100.0 | 0.18 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (100), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0), I8 (0), E9 (0), I9 (0), E10 (0), I10 (0), E11 (0), I11 (0), E12 (0), I12 (0), E13 (0), I13 (0), E14 (0), I14 (0), E15 (0), I15 (0), E16 (0), I16 (0), E17 (0), I17 (0), E18 (0), I18 (0), E19 (0), I19 (0), E20 (0), I20 (0), E21 (0), I21 (0), E22 (0), I22 (0), E23 (0), I23 (0), E24 (0), I24 (0), E25 (0), I25 (0), E26 (0), I26 (0), E27 (0) | 0.0 |
- 2 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 25121 | chr1 | 21978666 | 21979995 | wgEncodeRegDnaseClusteredV2 | 29 | score:753, srow:17362 |
| 25121 | chr1 | 21977741 | 21978818 | wgEncodeRegTfbsClusteredV3 | RBBP5 | score:219, expCount:1, expNums:33, expScores:207, srow:56680 |
| 25121 | chr1 | 21978329 | 21979312 | wgEncodeRegTfbsClusteredV3 | EZH2 | score:404, expCount:4, expNums:21,38,40,43, expScores:378,257,258,404, srow:56701 |
| 25121 | chr1 | 21978614 | 21978909 | wgEncodeRegTfbsClusteredV3 | RCOR1 | score:237, expCount:1, expNums:492, expScores:237, srow:56703 |
| 25121 | chr1 | 21978660 | 21978983 | wgEncodeRegTfbsClusteredV3 | CEBPB | score:187, expCount:2, expNums:212,477, expScores:168,187, srow:56704 |
| 25121 | chr1 | 21978875 | 21979394 | wgEncodeRegTfbsClusteredV3 | SIN3A | score:188, expCount:1, expNums:357, expScores:188, srow:56705 |
| 25121 | chr1 | 21978102 | 21978764 | cpgIsland | CpG: 60 | length:663, cpgNum:60, gcNum:357, perCpg:18.1, perGc:53.8, obsExp:1.28, srow:606 |
| 25121 | chr1 | 21978765 | 21980764 | cpgShore | — | values(x.gr)[overs$srow, ]:1064 |
- 8 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 25121 | cellmeth_recounts | PrEC: 34, LNCaP: 234, DU145: 5 | 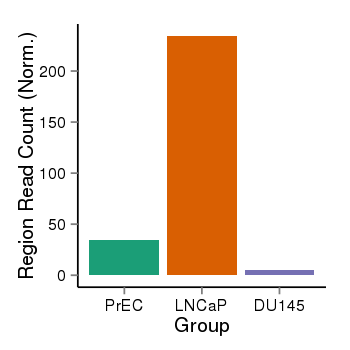 |
| 25121 | cellmeth_diffreps_PrECvsLNCaP | A=53, B=228, Length=680, Event=Up, log2FC=2.1, padj=0 | 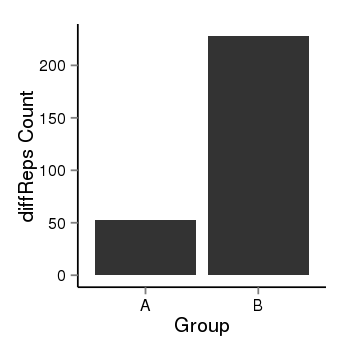 |
| 25121 | cellmeth_diffreps_PrECvsDU145 | A=53, B=4, Length=400, Event=Down, log2FC=-3.73, padj=1.90550538735206e-10 | 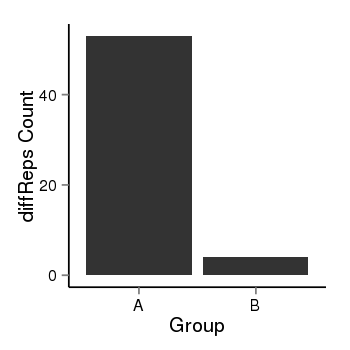 |
| 25121 | cellmeth_diffreps_LNCaPvsDU145 | A=243.74, B=3.74, Length=700, Event=Down, log2FC=-6.03, padj=0 | 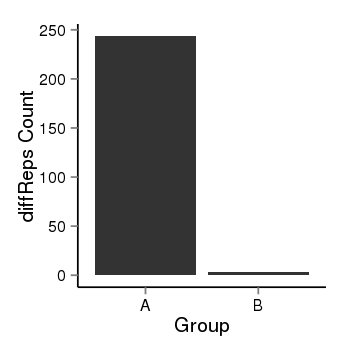 |
| 25121 | cellexp | id=ENSG00000076864, name=RAP1GAP, PrEC=105, str=98, LNCaP=1127, str=776, DU145=673, str=413 | 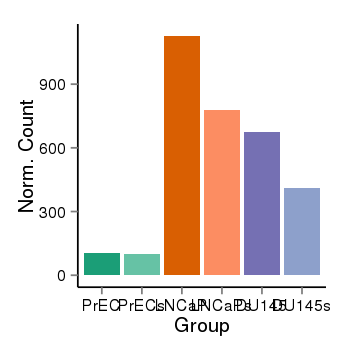 |
| 25121 | tcgameth | site:cg01723876, B=0.29, L=0.5, H=0.44 | 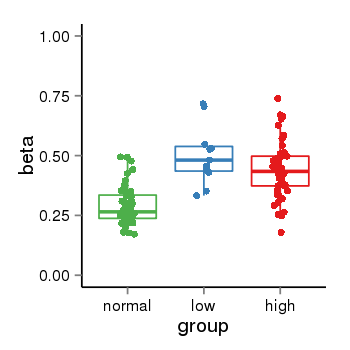 |
| 25121 | tcgameth | site:cg14515812, B=0.5, L=0.72, H=0.67 | 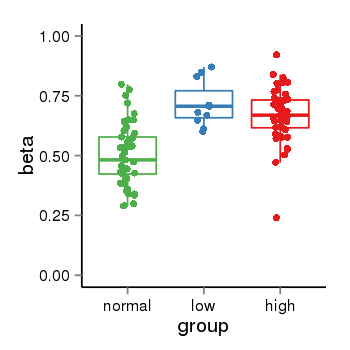 |
| 25121 | tcgaexp | gene=RAP1GAP, entrez=5909, pos=chr1:21922708-21995856(-), B=3563, L=8306, M=8429, H=8704 | 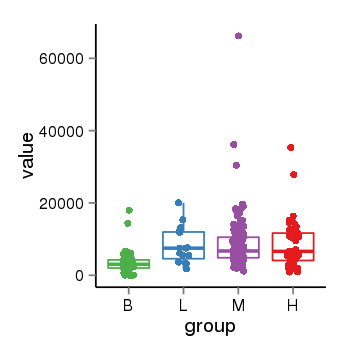 |
| 25121 | tcgaexp | gene=SRSF10, entrez=10772, pos=chr1:36273-24306953(-), B=3487, L=3388, M=3432, H=4019 | 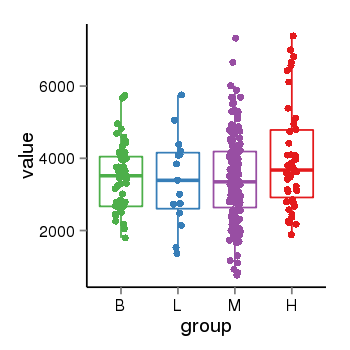 |
| 25121 | tcgaexp | gene=LOC100133331, entrez=100133331, pos=chr1:322037-180753553(-), B=268, L=328, M=271, H=305 | 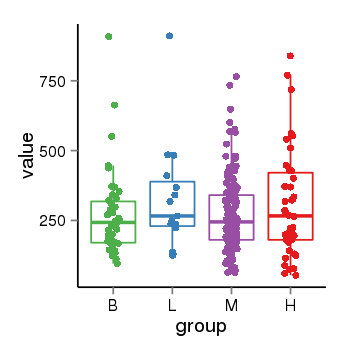 |