DMR 26235 chr2:65300801-65301000 (200bp)
| DMR Stats | |
|---|---|
| Position | chr2:65300801-65301000 (View on UCSC) |
| Width | 200bp |
| Genomic Context | 3' end |
| Nearest Genes | CEP68 (0bp away) |
| ANODEV p-value | 0.000365898327713 |
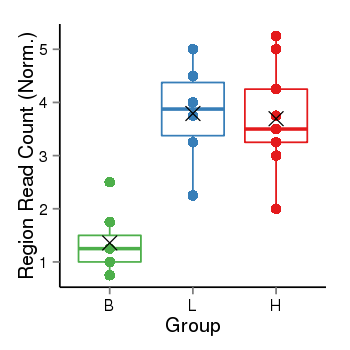
| Frequencies | Benign: 0.0 (0.0%), Low: 5.0 (83.33%), High: 7.0 (77.78 %) |
| Frequent? |
Region Count Boxplot
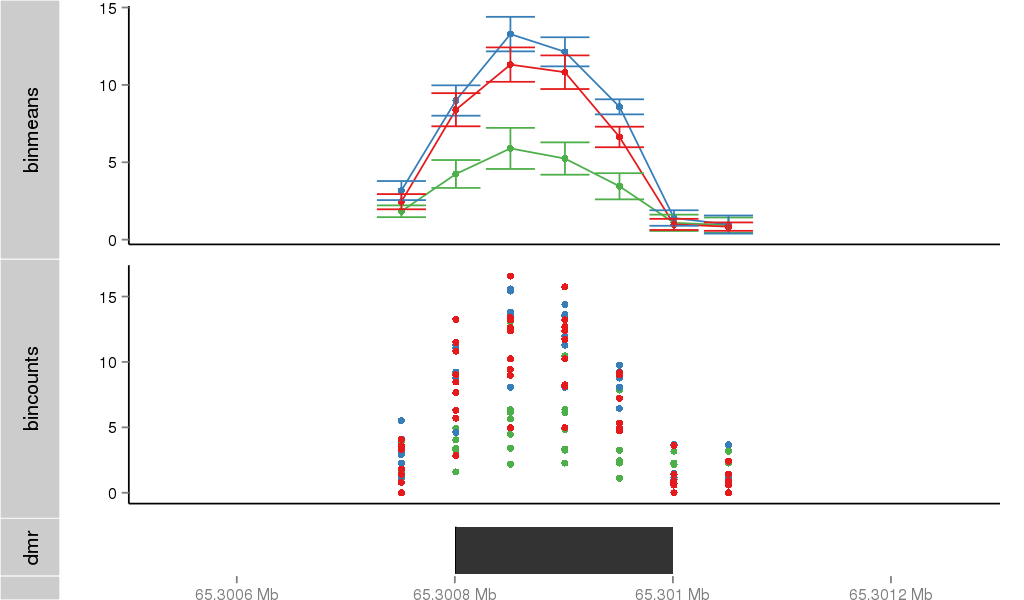
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 26235 | CEP68 | 16425 | uc002sdj.2 | chr2 | 65283495 | 65301312 | + | 200 | 100.0 | 0.14 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (100) | 100.0 |
| 26235 | CEP68 | 16425 | uc002sdk.4 | chr2 | 65283495 | 65314142 | + | 200 | 100.0 | 0.39 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (100), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0) | 0.0 |
| 26235 | CEP68 | 16425 | uc002sdl.4 | chr2 | 65283495 | 65314142 | + | 200 | 100.0 | 0.11 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (100), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0) | 0.0 |
| 26235 | CEP68 | 16425 | uc010yqb.1 | chr2 | 65283495 | 65309843 | + | 200 | 100.0 | 0.11 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (100), E4 (0), I4 (0), E5 (0) | 0.0 |
| 26235 | CEP68 | 16425 | uc010yqc.2 | chr2 | 65283550 | 65314142 | + | 200 | 100.0 | 0.14 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (100), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0) | 0.0 |
| 26235 | CEP68 | 16425 | uc010yqd.1 | chr2 | 65296533 | 65301622 | + | 200 | 100.0 | 0.11 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (100), E3 (0) | 100.0 |
- 6 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 26235 | chr2 | 65300686 | 65300955 | wgEncodeRegDnaseClusteredV2 | 3 | score:176, srow:625241 |
- 1 features
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 26235 | cellmeth_recounts | PrEC: 1, LNCaP: 0, DU145: 1 | 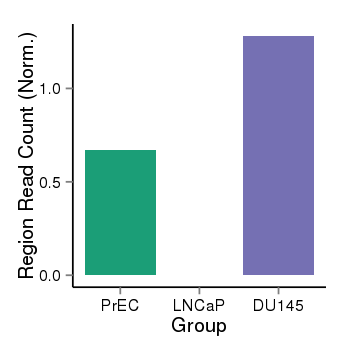 |
| 26235 | cellexp | id=ENSG00000011523, name=CEP68, PrEC=1242, str=851, LNCaP=994, str=808, DU145=1706, str=1413 | 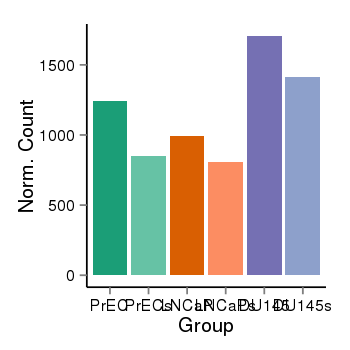 |
| 26235 | cellexp | id=ENSG00000138069, name=RAB1A, PrEC=13740, str=16758, LNCaP=5067, str=6247, DU145=7770, str=7942 | 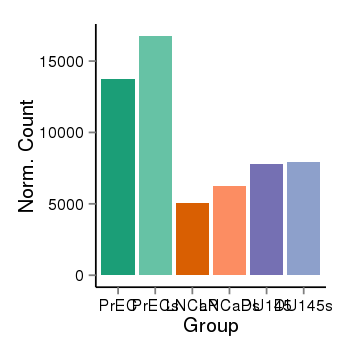 |
| 26235 | tcgaexp | gene=CEP68, entrez=23177, pos=chr2:65283495-65314142(+), B=2398, L=1459, M=1516, H=1476 | 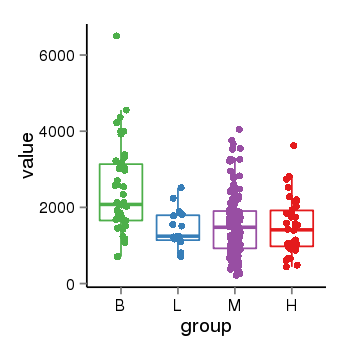 |