DMR 283 chr1:55246551-55246800 (250bp)
| DMR Stats | |
|---|---|
| Position | chr1:55246551-55246800 (View on UCSC) |
| Width | 250bp |
| Genomic Context | 3' end |
| Nearest Genes | TTC22 (0bp away) |
| ANODEV p-value | 0.0208538849247 |
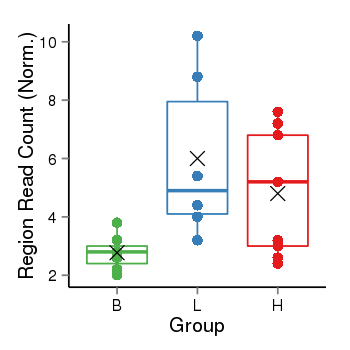
| Frequencies | Benign: 2.0 (28.57%), Low: 6.0 (100.0%), High: 6.0 (66.67 %) |
| Frequent? |
Region Count Boxplot
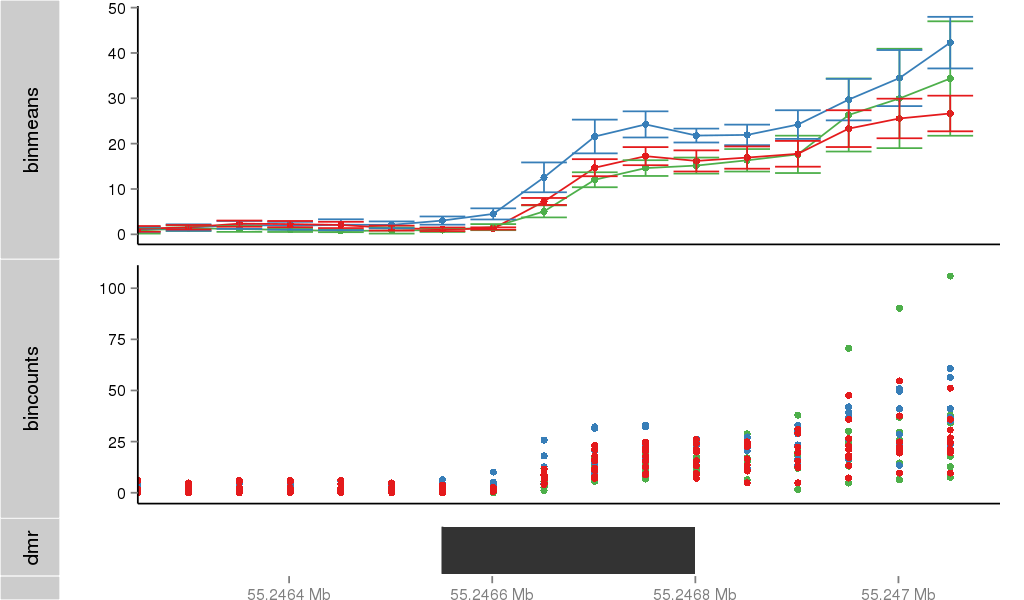
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 283 | TTC22 | 924 | uc009vzt.1 | chr1 | 55246752 | 55266941 | - | 49 | 19.6 | 0.3 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (19.6) | 100.0 |
- 1 genes
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 283 | chr1 | 55246766 | 55246935 | wgEncodeRegDnaseClusteredV2 | 4 | score:786, srow:41707 |
| 283 | chr1 | 55246632 | 55246919 | wgEncodeRegTfbsClusteredV3 | CEBPB | score:587, expCount:6, expNums:277,343,425,426,462,477, expScores:253,553,145,382,587,248, srow:147805 |
| 283 | chr1 | 55246700 | 55247742 | wgEncodeRegTfbsClusteredV3 | EZH2 | score:348, expCount:2, expNums:21,38, expScores:348,219, srow:147806 |
| 283 | chr1 | 55246743 | 55247519 | cpgIsland | CpG: 88 | length:777, cpgNum:88, gcNum:560, perCpg:22.7, perGc:72.1, obsExp:0.88, srow:1158 |
| 283 | chr1 | 55244743 | 55246742 | cpgShore | — | values(x.gr)[overs$srow, ]:2112 |
- 5 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 283 | cellmeth_recounts | PrEC: 13, LNCaP: 68, DU145: 9 | 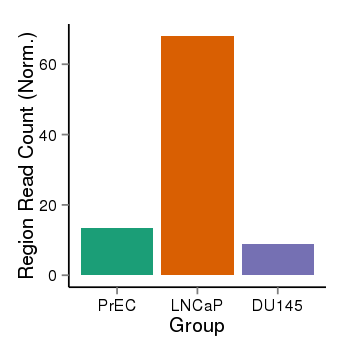 |
| 283 | cellmeth_diffreps_PrECvsLNCaP | A=19, B=53, Length=200, Event=Up, log2FC=1.48, padj=0.00157949948552458 | 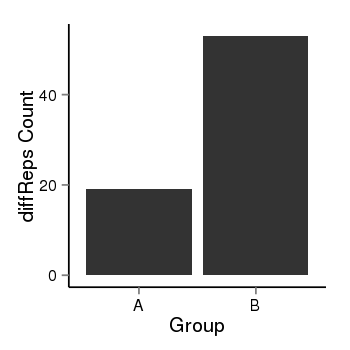 |
| 283 | cellmeth_diffreps_PrECvsDU145 | A=780, B=28, Length=1000, Event=Down, log2FC=-4.8, padj=0 | 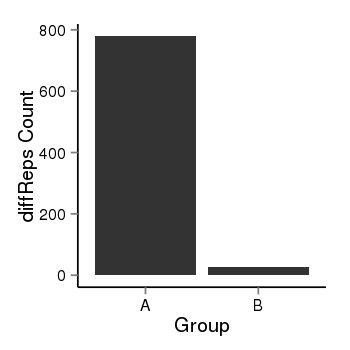 |
| 283 | cellmeth_diffreps_LNCaPvsDU145 | A=730.16, B=27.13, Length=1060, Event=Down, log2FC=-4.75, padj=0 | 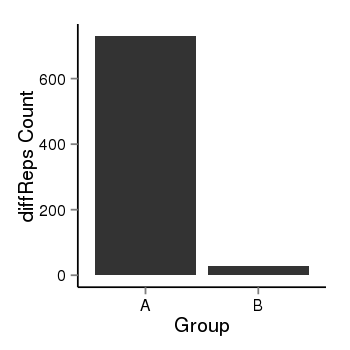 |
| 283 | cellexp | id=ENSG00000006555, name=TTC22, PrEC=998, str=553, LNCaP=8, str=0, DU145=535, str=337 | 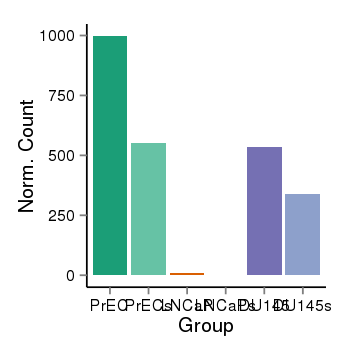 |
| 283 | tcgaexp | gene=TTC22, entrez=55001, pos=chr1:55246752-55266941(-), B=297, L=172, M=195, H=147 | 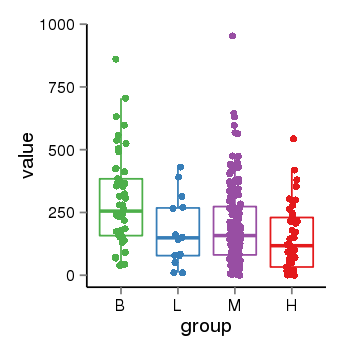 |
| 283 | tcgaexp | gene=LOC100133331, entrez=100133331, pos=chr1:322037-180753553(-), B=268, L=328, M=271, H=305 | 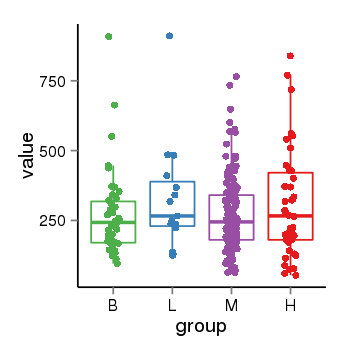 |