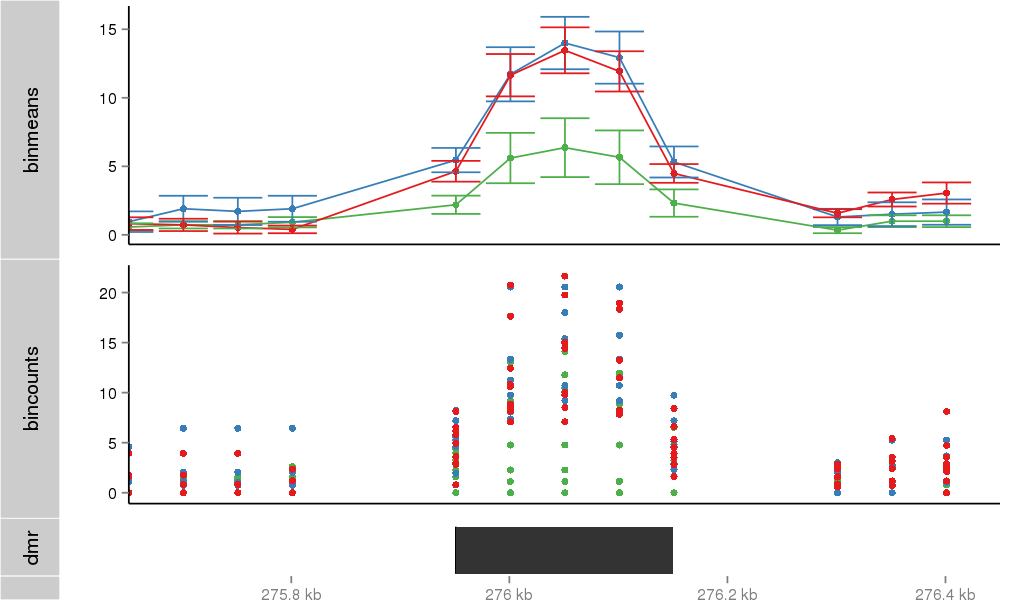
DMR 31967 chr12:275951-276150 (200bp)
| DMR Stats | |
|---|---|
| Position | chr12:275951-276150 (View on UCSC) |
| Width | 200bp |
| Genomic Context | intron |
| Nearest Genes | IQSEC3 (0bp away) |
| ANODEV p-value | 0.012972349828 |
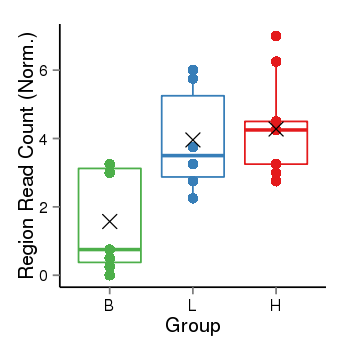
| Frequencies | Benign: 2.0 (28.57%), Low: 4.0 (66.67%), High: 7.0 (77.78 %) |
| Frequent? |
Region Count Boxplot
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 31967 | IQSEC3 | 5882 | uc001qhu.1 | chr12 | 186542 | 280494 | + | 200 | 100.0 | 1.77 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0), I8 (0), E9 (0), I9 (0), E10 (0), I10 (100), E11 (0), I11 (0), E12 (0), I12 (0), E13 (0) | 0.0 |
| 31967 | IQSEC3 | 5882 | uc001qhw.2 | chr12 | 176049 | 287625 | + | 200 | 100.0 | 4.25 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0), I8 (0), E9 (0), I9 (0), E10 (0), I10 (0), E11 (0), I11 (100), E12 (0), I12 (0), E13 (0), I13 (0), E14 (0) | 0.0 |
- 2 geness
Feature Overlaps
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 31967 | cellmeth_recounts | PrEC: 7, LNCaP: 4, DU145: 0 | 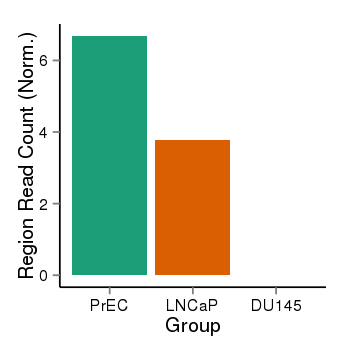 |
| 31967 | cellexp | id=ENSG00000120645, name=IQSEC3, PrEC=1, str=4, LNCaP=5, str=2, DU145=3, str=3 | 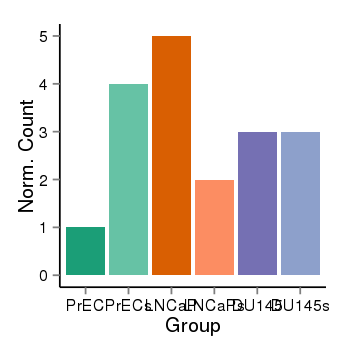 |
| 31967 | cellexp | id=ENSG00000256540, name=RP11-598F7.6, PrEC=0, str=0, LNCaP=0, str=0, DU145=10, str=7 | 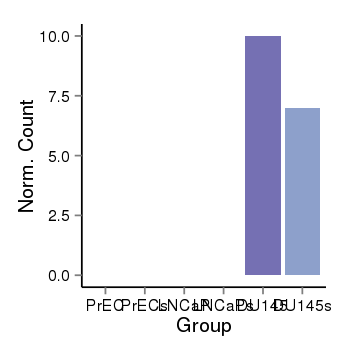 |
| 31967 | tcgaexp | gene=ZNF26, entrez=7574, pos=chr12:93510-133589154(+), B=419, L=392, M=412, H=512 | 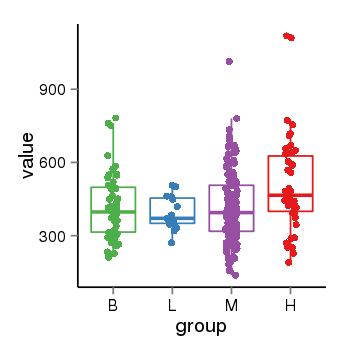 |
| 31967 | tcgaexp | gene=ZNF84, entrez=7637, pos=chr12:42780-133639885(+), B=1626, L=1649, M=1716, H=2045 | 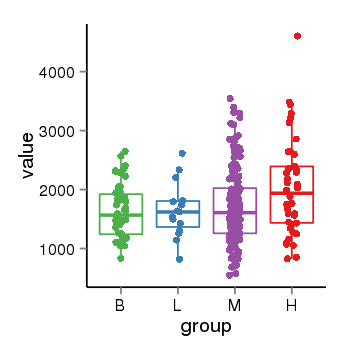 |
| 31967 | tcgaexp | gene=IQSEC3, entrez=440073, pos=chr12:176049-287625(+), B=193, L=82, M=63, H=58 | 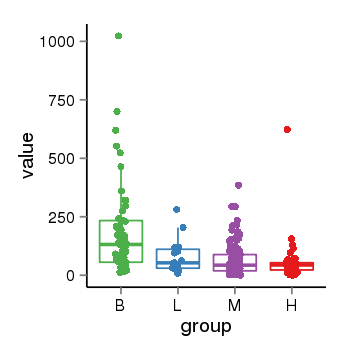 |