DMR 33875 chr17:753151-753350 (200bp)
| DMR Stats | |
|---|---|
| Position | chr17:753151-753350 (View on UCSC) |
| Width | 200bp |
| Genomic Context | intron |
| Nearest Genes | NXN (0bp away) |
| ANODEV p-value | 0.00156077832716 |
| Frequencies | Benign: 1.0 (14.29%), Low: 5.0 (83.33%), High: 9.0 (100.0 %) |
| Frequent? |
Region Count Boxplot

Region Overview

Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 33875 | NXN | 11889 | uc002fsa.3 | chr17 | 702553 | 882998 | - | 200 | 100.0 | 0.04 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0) | 0.0 |
| 33875 | NXN | 11890 | uc002fsb.2 | chr17 | 722368 | 800254 | - | 200 | 100.0 | 0.04 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0) | 0.0 |
| 33875 | NXN | 11889 | uc010vqe.2 | chr17 | 702553 | 767351 | - | 200 | 100.0 | 0.04 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0) | 0.0 |
- 3 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 33875 | chr17 | 752746 | 753395 | wgEncodeRegDnaseClusteredV2 | 10 | score:185, srow:471476 |
| 33875 | chr17 | 753311 | 753320 | tfbsConsSites | V$FOXO1_01 | score:970, zScore:2, srow:2057510 |
| 33875 | chr17 | 753312 | 753322 | tfbsConsSites | V$FOXO4_01 | score:953, zScore:2.4, srow:2057511 |
| 33875 | chr17 | 753219 | 753225 | phastConsElements100way | lod=22 | score:300, srow:3520649 |
| 33875 | chr17 | 753263 | 753267 | phastConsElements100way | lod=13 | score:247, srow:3520650 |
| 33875 | chr17 | 753285 | 753305 | phastConsElements100way | lod=31 | score:333, srow:3520651 |
| 33875 | chr17 | 753312 | 753318 | phastConsElements100way | lod=21 | score:295, srow:3520652 |
| 33875 | chr17 | 753331 | 753336 | phastConsElements100way | lod=15 | score:262, srow:3520653 |
- 8 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 33875 | cellmeth_recounts | PrEC: 9, LNCaP: 6, DU145: 8 |  |
| 33875 | cellexp | id=ENSG00000167693, name=NXN, PrEC=14170, str=7994, LNCaP=1525, str=1905, DU145=11809, str=9379 |  |
| 33875 | tcgaexp | gene=NSF, entrez=4905, pos=chr17:266212-44834828(+), B=3355, L=5678, M=5410, H=5700 | 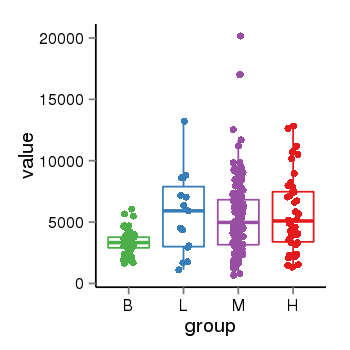 |
| 33875 | tcgaexp | gene=PLEKHM1, entrez=9842, pos=chr17:128328-43568146(-), B=1993, L=1631, M=1589, H=1418 | 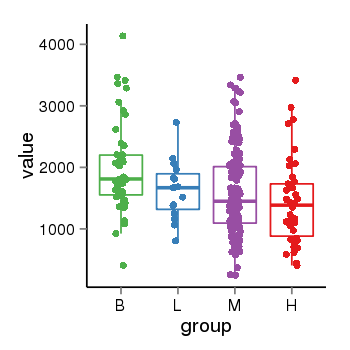 |
| 33875 | tcgaexp | gene=LRRC37A, entrez=9884, pos=chr17:415627-44415160(+), B=924, L=896, M=971, H=940 | 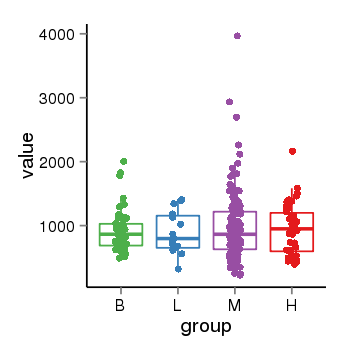 |
| 33875 | tcgaexp | gene=ARL17A, entrez=51326, pos=chr17:195721-44657088(-), B=200, L=195, M=210, H=230 | 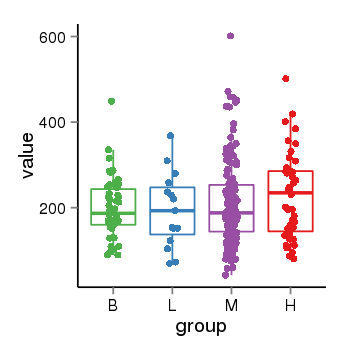 |
| 33875 | tcgaexp | gene=LRRC37A4P, entrez=55073, pos=chr17:197706-43597889(-), B=1015, L=1512, M=1396, H=1450 | 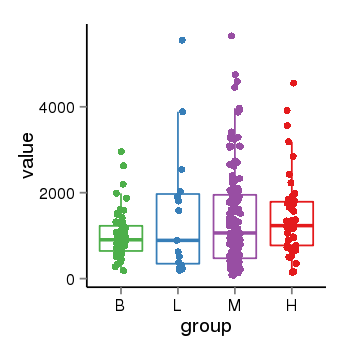 |
| 33875 | tcgaexp | gene=NXN, entrez=64359, pos=chr17:702553-882998(-), B=5704, L=5393, M=4923, H=3951 |  |
| 33875 | tcgaexp | gene=ARHGAP27, entrez=201176, pos=chr17:86404-43511112(-), B=1570, L=1264, M=1273, H=1486 | 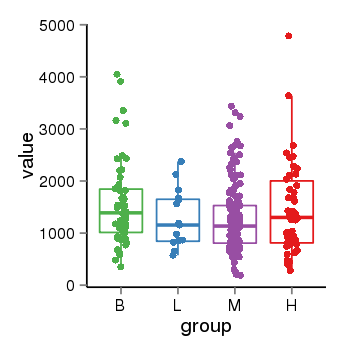 |
| 33875 | tcgaexp | gene=KANSL1, entrez=284058, pos=chr17:563498-44302740(-), B=3141, L=2594, M=2797, H=3017 | 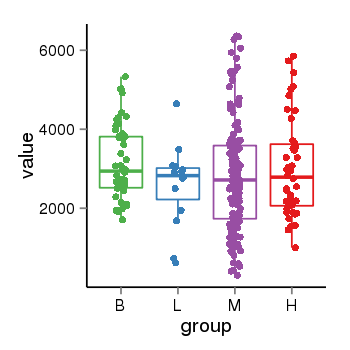 |
| 33875 | tcgaexp | gene=LRRC37A2, entrez=474170, pos=chr17:197706-44633014(+), B=351, L=310, M=361, H=377 | 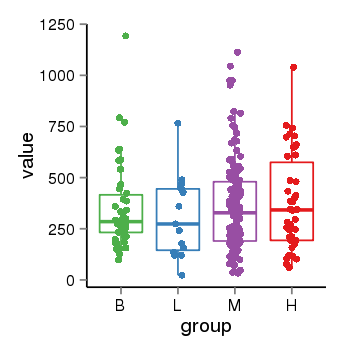 |