DMR 33881 chr17:807051-807700 (650bp)
| DMR Stats | |
|---|---|
| Position | chr17:807051-807700 (View on UCSC) |
| Width | 650bp |
| Genomic Context | intron |
| Nearest Genes | NXN (0bp away) |
| ANODEV p-value | 6.32444760828e-07 |
| Frequencies | Benign: 0.0 (0.0%), Low: 6.0 (100.0%), High: 9.0 (100.0 %) |
| Frequent? |
Region Count Boxplot

Region Overview

Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 33881 | NXN | 11889 | uc002fsa.3 | chr17 | 702553 | 882998 | - | 650 | 100.0 | 0.05 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0) | 0.0 |
- 1 genes
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 33881 | chr17 | 806986 | 807135 | wgEncodeRegDnaseClusteredV2 | 3 | score:314, srow:471532 |
| 33881 | chr17 | 807146 | 807770 | wgEncodeRegDnaseClusteredV2 | 71 | score:1000, srow:471533 |
| 33881 | chr17 | 806718 | 807719 | wgEncodeRegTfbsClusteredV3 | POLR2A | score:269, expCount:2, expNums:41,563, expScores:153,269, srow:1607290 |
| 33881 | chr17 | 807108 | 807507 | wgEncodeRegTfbsClusteredV3 | FOS | score:527, expCount:5, expNums:457,556,557,558,559, expScores:159,527,455,398,320, srow:1607291 |
| 33881 | chr17 | 807229 | 807538 | wgEncodeRegTfbsClusteredV3 | MYC | score:192, expCount:1, expNums:560, expScores:192, srow:1607292 |
| 33881 | chr17 | 807340 | 807362 | tfbsConsSites | V$OCT1_04 | score:798, zScore:1.78, srow:2057585 |
| 33881 | chr17 | 807351 | 807364 | tfbsConsSites | V$CHX10_01 | score:828, zScore:2.13, srow:2057586 |
| 33881 | chr17 | 807379 | 807393 | tfbsConsSites | V$SRF_C | score:783, zScore:1.66, srow:2057587 |
| 33881 | chr17 | 807383 | 807391 | tfbsConsSites | V$MSX1_01 | score:904, zScore:1.86, srow:2057588 |
| 33881 | chr17 | 807695 | 807700 | tfbsConsSites | V$AML1_01 | score:1000, zScore:1.78, srow:2057589 |
| 33881 | chr17 | 807084 | 807086 | phastConsElements100way | lod=12 | score:240, srow:3520840 |
| 33881 | chr17 | 807210 | 807228 | phastConsElements100way | lod=58 | score:395, srow:3520841 |
| 33881 | chr17 | 807264 | 807282 | phastConsElements100way | lod=97 | score:446, srow:3520842 |
| 33881 | chr17 | 807293 | 807313 | phastConsElements100way | lod=72 | score:417, srow:3520843 |
| 33881 | chr17 | 807316 | 807327 | phastConsElements100way | lod=90 | score:439, srow:3520844 |
| 33881 | chr17 | 807337 | 807370 | phastConsElements100way | lod=189 | score:512, srow:3520845 |
| 33881 | chr17 | 807373 | 807408 | phastConsElements100way | lod=105 | score:454, srow:3520846 |
| 33881 | chr17 | 807423 | 807428 | phastConsElements100way | lod=18 | score:280, srow:3520847 |
| 33881 | chr17 | 807607 | 807611 | phastConsElements100way | lod=15 | score:262, srow:3520848 |
- 19 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 33881 | cellmeth_recounts | PrEC: 0, LNCaP: 6, DU145: 20 |  |
| 33881 | cellexp | id=ENSG00000167693, name=NXN, PrEC=14170, str=7994, LNCaP=1525, str=1905, DU145=11809, str=9379 |  |
| 33881 | tcgameth | site:cg04954757, B=0.53, L=0.78, H=0.75 |  |
| 33881 | tcgameth | site:cg10194536, B=0.34, L=0.64, H=0.61 |  |
| 33881 | tcgaexp | gene=MAPT, entrez=4137, pos=chr17:762281-44105699(+), B=1933, L=2382, M=2487, H=2583 | 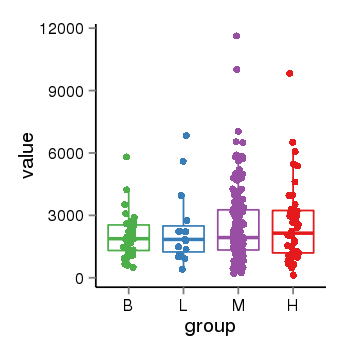 |
| 33881 | tcgaexp | gene=NSF, entrez=4905, pos=chr17:266212-44834828(+), B=3355, L=5678, M=5410, H=5700 | 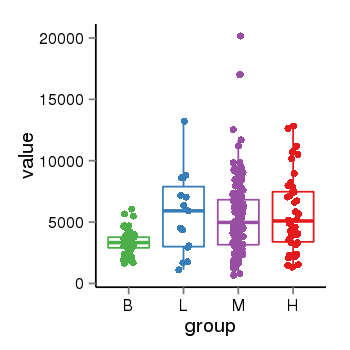 |
| 33881 | tcgaexp | gene=PLEKHM1, entrez=9842, pos=chr17:128328-43568146(-), B=1993, L=1631, M=1589, H=1418 | 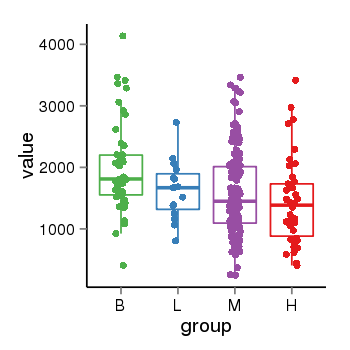 |
| 33881 | tcgaexp | gene=LRRC37A, entrez=9884, pos=chr17:415627-44415160(+), B=924, L=896, M=971, H=940 | 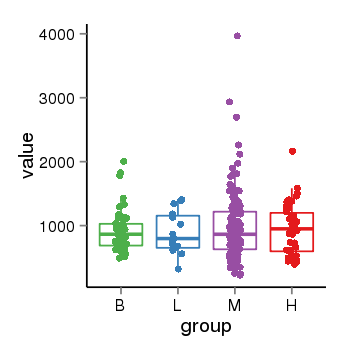 |
| 33881 | tcgaexp | gene=ARL17A, entrez=51326, pos=chr17:195721-44657088(-), B=200, L=195, M=210, H=230 | 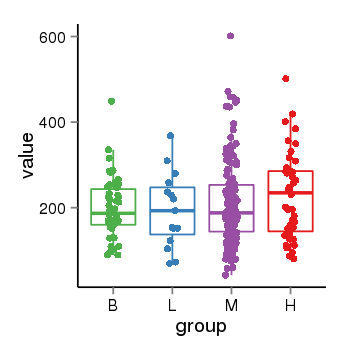 |
| 33881 | tcgaexp | gene=LRRC37A4P, entrez=55073, pos=chr17:197706-43597889(-), B=1015, L=1512, M=1396, H=1450 | 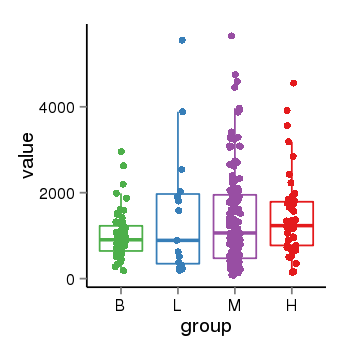 |
| 33881 | tcgaexp | gene=NXN, entrez=64359, pos=chr17:702553-882998(-), B=5704, L=5393, M=4923, H=3951 |  |
| 33881 | tcgaexp | gene=ARHGAP27, entrez=201176, pos=chr17:86404-43511112(-), B=1570, L=1264, M=1273, H=1486 | 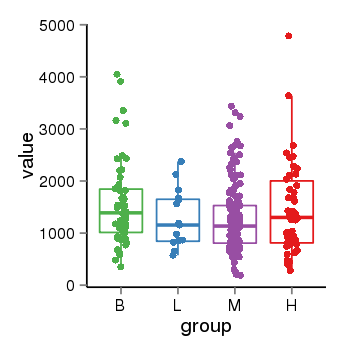 |
| 33881 | tcgaexp | gene=STH, entrez=246744, pos=chr17:790689-44077060(+), B=0, L=0, M=0, H=0 | 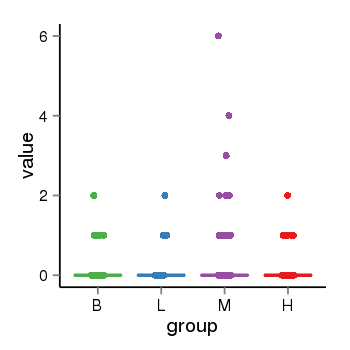 |
| 33881 | tcgaexp | gene=KANSL1, entrez=284058, pos=chr17:563498-44302740(-), B=3141, L=2594, M=2797, H=3017 | 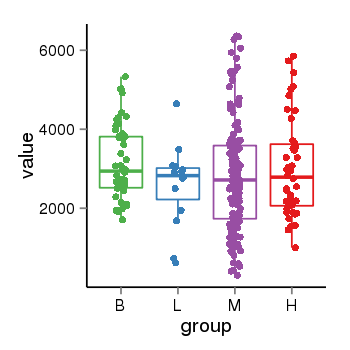 |
| 33881 | tcgaexp | gene=LRRC37A2, entrez=474170, pos=chr17:197706-44633014(+), B=351, L=310, M=361, H=377 | 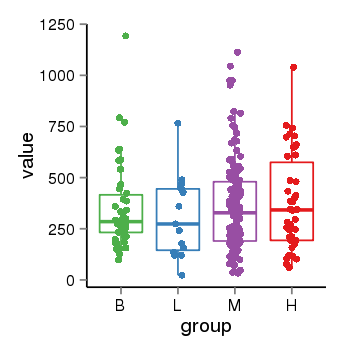 |