DMR 36533 chr1:2520251-2520400 (150bp)
| DMR Stats | |
|---|---|
| Position | chr1:2520251-2520400 (View on UCSC) |
| Width | 150bp |
| Genomic Context | exon |
| Nearest Genes | FAM213B (0bp away) |
| ANODEV p-value | 0.0048688172153 |
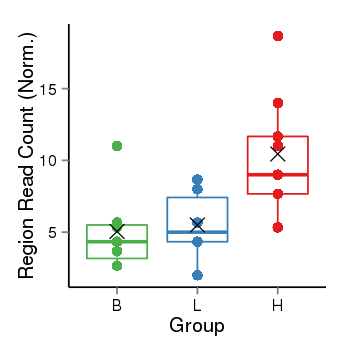
| Frequencies | Benign: 5.0 (71.43%), Low: 5.0 (83.33%), High: 9.0 (100.0 %) |
| Frequent? |
Region Count Boxplot
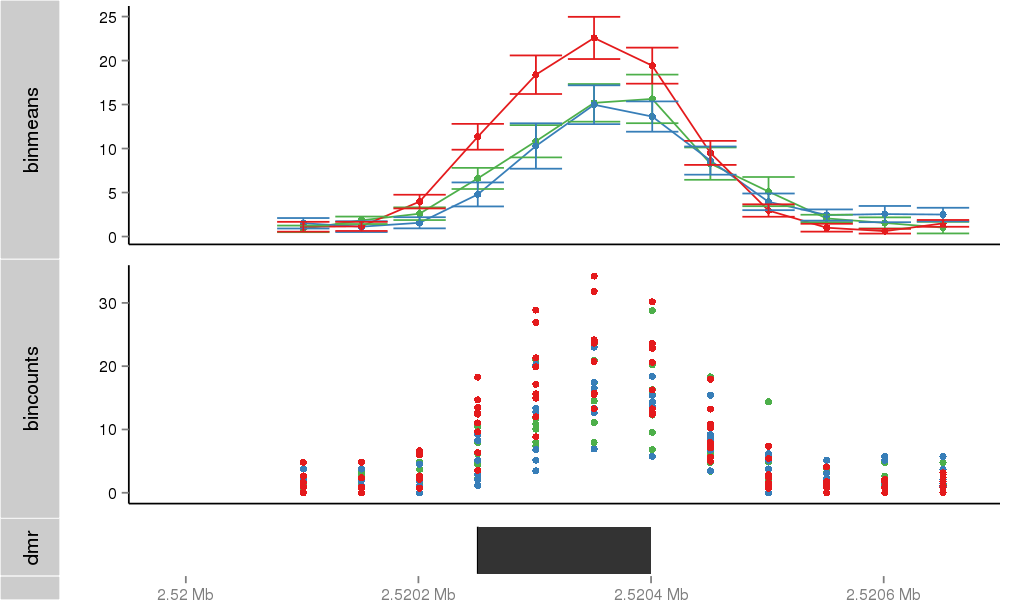
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 36533 | FAM213B | 111 | uc001aju.3 | chr1 | 2518189 | 2522908 | + | 150 | 100.0 | 0.05 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (73.33), E6 (26.67), I6 (0), E7 (0) | 0.0 |
| 36533 | FAM213B | 111 | uc001ajv.2 | chr1 | 2518189 | 2522908 | + | 150 | 100.0 | 0.72 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (73.33), E6 (26.67), I6 (0), E7 (0) | 0.0 |
| 36533 | FAM213B | 111 | uc001ajw.2 | chr1 | 2518189 | 2522908 | + | 150 | 100.0 | 0.08 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (94.67), E6 (5.33), I6 (0), E7 (0) | 0.0 |
| 36533 | FAM213B | 111 | uc001ajx.2 | chr1 | 2518517 | 2522908 | + | 150 | 100.0 | 0.08 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (94.67), E5 (5.33), I5 (0), E6 (0) | 0.0 |
| 36533 | FAM213B | 111 | uc010nzd.2 | chr1 | 2518189 | 2522908 | + | 150 | 100.0 | 0.64 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (94.67), E6 (5.33), I6 (0), E7 (0) | 0.0 |
| 36533 | FAM213B | 111 | uc010nze.2 | chr1 | 2518189 | 2522908 | + | 150 | 100.0 | 0.64 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (94.67), E5 (5.33), I5 (0), E6 (0) | 0.0 |
| 36533 | FAM213B | 111 | uc010nzf.2 | chr1 | 2518189 | 2522908 | + | 150 | 100.0 | 0.64 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (96.67), E5 (3.33), I5 (0), E6 (0) | 0.0 |
| 36533 | FAM213B | 111 | uc021oew.1 | chr1 | 2517899 | 2522908 | + | 150 | 100.0 | 0.64 | 1 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (94.67), E6 (5.33), I6 (0), E7 (0) | 0.0 |
| 36533 | FAM213B | 111 | uc021oex.1 | chr1 | 2517899 | 2522908 | + | 150 | 100.0 | 0.49 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (73.33), E6 (26.67), I6 (0), E7 (0) | 0.0 |
- 9 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 36533 | chr1 | 2520166 | 2520695 | wgEncodeRegDnaseClusteredV2 | 6 | score:356, srow:1748 |
| 36533 | chr1 | 2520154 | 2520749 | wgEncodeRegTfbsClusteredV3 | ZNF263 | score:259, expCount:1, expNums:368, expScores:259, srow:8347 |
| 36533 | chr1 | 2520353 | 2520368 | tfbsConsSites | V$TAL1BETAITF2_01 | score:846, zScore:1.73, srow:4371 |
| 36533 | chr1 | 2520382 | 2520382 | cosmic | COSM40320 | srow:1960 |
| 36533 | chr1 | 2520383 | 2520383 | cosmic | COSM1341236 | srow:1961 |
| 36533 | chr1 | 2520386 | 2520386 | cosmic | COSM907454 | srow:1962 |
| 36533 | chr1 | 2520399 | 2520399 | cosmic | COSM178376 | srow:1963 |
| 36533 | chr1 | 2520338 | 2520340 | phastConsElements100way | lod=18 | score:280, srow:11424 |
| 36533 | chr1 | 2520359 | 2520367 | phastConsElements100way | lod=70 | score:414, srow:11425 |
| 36533 | chr1 | 2520372 | 2520379 | phastConsElements100way | lod=35 | score:345, srow:11426 |
| 36533 | chr1 | 2520381 | 2520385 | phastConsElements100way | lod=33 | score:340, srow:11427 |
| 36533 | chr1 | 2520390 | 2520397 | phastConsElements100way | lod=59 | score:397, srow:11428 |
| 36533 | chr1 | 2520399 | 2520400 | phastConsElements100way | lod=17 | score:274, srow:11429 |
| 36533 | chr1 | 2518893 | 2520892 | cpgShore | — | values(x.gr)[overs$srow, ]:347 |
- 14 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 36533 | cellmeth_recounts | PrEC: 33, LNCaP: 19, DU145: 4 | 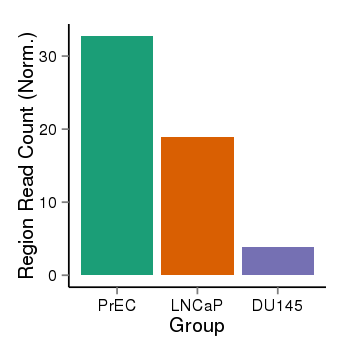 |
| 36533 | cellmeth_diffreps_PrECvsLNCaP | A=58, B=19, Length=300, Event=Down, log2FC=-1.61, padj=0.000235448342019214 | 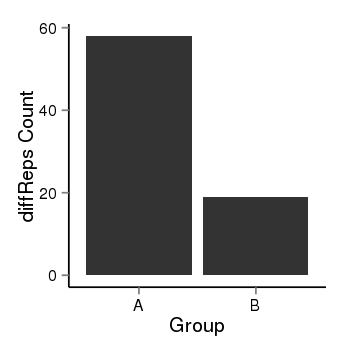 |
| 36533 | cellmeth_diffreps_PrECvsDU145 | A=58, B=6, Length=460, Event=Down, log2FC=-3.27, padj=3.36978979971677e-10 |  |
| 36533 | cellexp | id=ENSG00000157870, name=FAM213B, PrEC=3023, str=1367, LNCaP=3832, str=3142, DU145=1558, str=1253 | 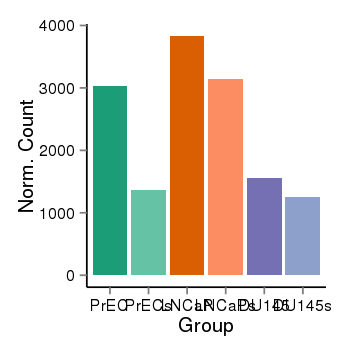 |
| 36533 | tcgaexp | gene=SRSF10, entrez=10772, pos=chr1:36273-24306953(-), B=3487, L=3388, M=3432, H=4019 | 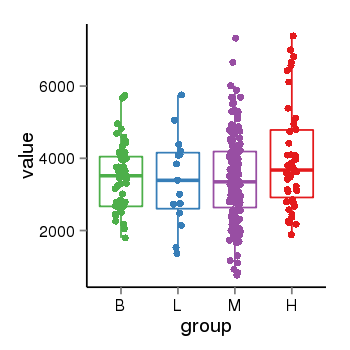 |
| 36533 | tcgaexp | gene=FAM213B, entrez=127281, pos=chr1:2517899-2522908(+), B=2251, L=2968, M=2657, H=2580 | 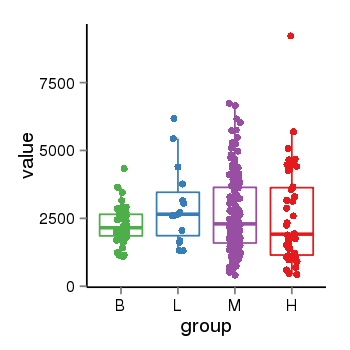 |
| 36533 | tcgaexp | gene=LOC100133331, entrez=100133331, pos=chr1:322037-180753553(-), B=268, L=328, M=271, H=305 | 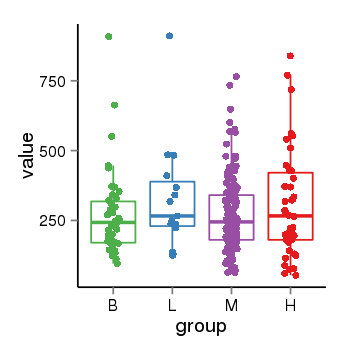 |