DMR 36611 chr1:15392851-15393000 (150bp)
| DMR Stats | |
|---|---|
| Position | chr1:15392851-15393000 (View on UCSC) |
| Width | 150bp |
| Genomic Context | exon |
| Nearest Genes | KAZN (0bp away) |
| ANODEV p-value | 0.000624921913548 |
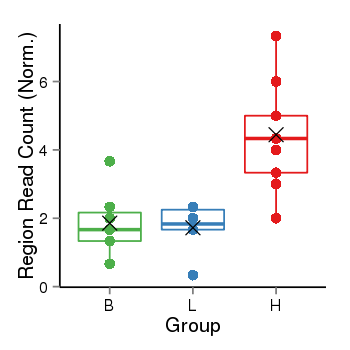
| Frequencies | Benign: 1.0 (14.29%), Low: 0.0 (0.0%), High: 7.0 (77.78 %) |
| Frequent? |
Region Count Boxplot
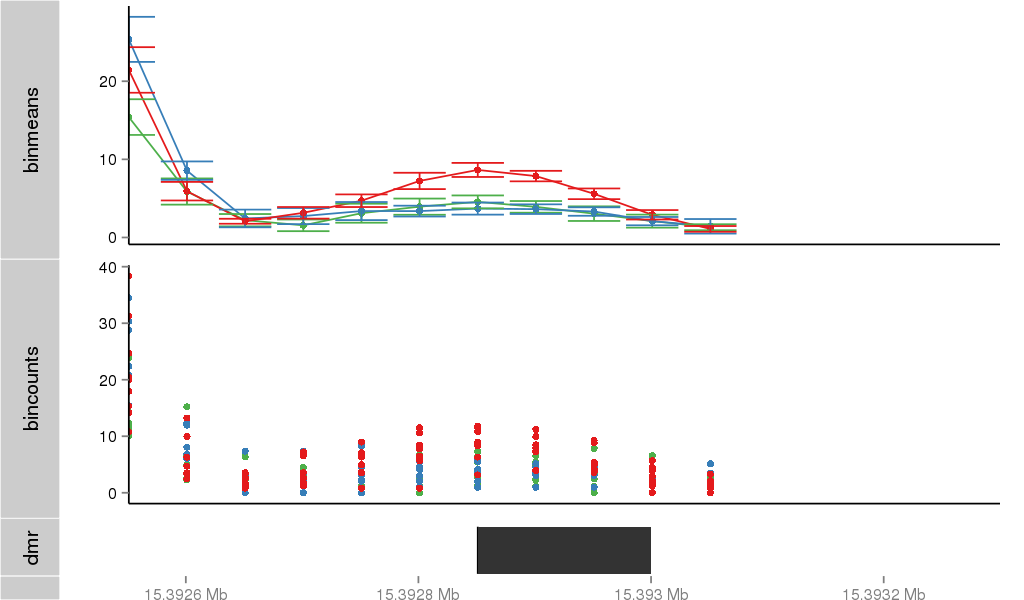
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 36611 | KAZN | 266 | uc001avm.4 | chr1 | 14925213 | 15444544 | + | 150 | 100.0 | 0.03 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0), I8 (100), E9 (0), I9 (0), E10 (0), I10 (0), E11 (0), I11 (0), E12 (0), I12 (0), E13 (0), I13 (0), E14 (0), I14 (0), E15 (0) | 0.0 |
| 36611 | KAZN | 266 | uc001avo.2 | chr1 | 15250625 | 15394651 | + | 150 | 100.0 | 0.04 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (100) | 0.0 |
| 36611 | KAZN | 266 | uc001avp.2 | chr1 | 15256296 | 15394651 | + | 150 | 100.0 | 0.26 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (100) | 0.0 |
| 36611 | KAZN | 266 | uc001avq.2 | chr1 | 15272415 | 15394651 | + | 150 | 100.0 | 0.13 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (100) | 0.0 |
| 36611 | KAZN | 266 | uc001avr.2 | chr1 | 15287180 | 15394651 | + | 150 | 100.0 | 0.19 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (100) | 0.0 |
| 36611 | KAZN | 266 | uc009vog.1 | chr1 | 14925213 | 15394651 | + | 150 | 100.0 | 0.19 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (100) | 0.0 |
- 6 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 36611 | chr1 | 15392706 | 15392950 | wgEncodeRegDnaseClusteredV2 | 5 | score:178, srow:11665 |
| 36611 | chr1 | 15392845 | 15392860 | tfbsConsSites | V$ARNT_01 | score:847, zScore:1.88, srow:31390 |
| 36611 | chr1 | 15392858 | 15392876 | tfbsConsSites | V$GR_Q6 | score:824, zScore:1.7, srow:31391 |
| 36611 | chr1 | 15392898 | 15392903 | tfbsConsSites | V$AML1_01 | score:1000, zScore:1.78, srow:31392 |
| 36611 | chr1 | 15392907 | 15392916 | tfbsConsSites | V$GATA1_01 | score:939, zScore:1.72, srow:31393 |
| 36611 | chr1 | 15392907 | 15392916 | tfbsConsSites | V$GATA2_01 | score:946, zScore:1.83, srow:31394 |
| 36611 | chr1 | 15392910 | 15392923 | tfbsConsSites | V$ATF_01 | score:889, zScore:2.79, srow:31395 |
| 36611 | chr1 | 15392985 | 15392997 | tfbsConsSites | V$RORA1_01 | score:876, zScore:1.84, srow:31396 |
| 36611 | chr1 | 15392998 | 15393019 | tfbsConsSites | V$MEF2_02 | score:777, zScore:1.7, srow:31397 |
| 36611 | chr1 | 15392857 | 15392870 | phastConsElements100way | lod=28 | score:323, srow:55998 |
| 36611 | chr1 | 15392877 | 15392879 | phastConsElements100way | lod=13 | score:247, srow:55999 |
| 36611 | chr1 | 15392883 | 15392886 | phastConsElements100way | lod=18 | score:280, srow:56000 |
| 36611 | chr1 | 15392888 | 15392897 | phastConsElements100way | lod=29 | score:327, srow:56001 |
| 36611 | chr1 | 15392919 | 15392924 | phastConsElements100way | lod=22 | score:300, srow:56002 |
| 36611 | chr1 | 15392976 | 15392978 | phastConsElements100way | lod=19 | score:285, srow:56003 |
| 36611 | chr1 | 15392992 | 15392994 | phastConsElements100way | lod=19 | score:285, srow:56004 |
- 16 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 36611 | cellmeth_recounts | PrEC: 6, LNCaP: 1, DU145: 3 | 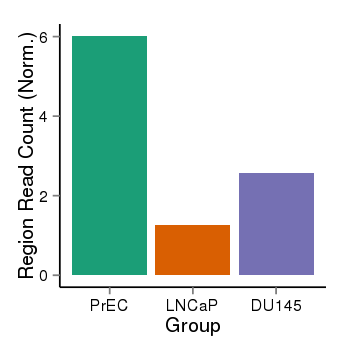 |
| 36611 | cellexp | id=ENSG00000189337, name=KAZN, PrEC=1257, str=890, LNCaP=399, str=388, DU145=394, str=284 | 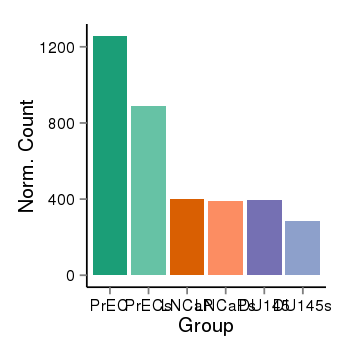 |
| 36611 | tcgameth | site:cg16552271, B=0.66, L=0.76, H=0.74 | 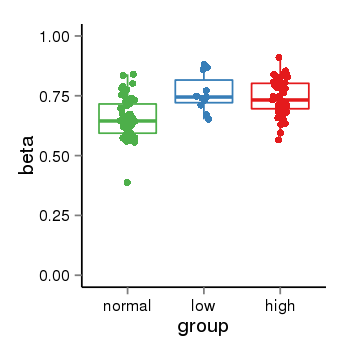 |
| 36611 | tcgaexp | gene=SRSF10, entrez=10772, pos=chr1:36273-24306953(-), B=3487, L=3388, M=3432, H=4019 | 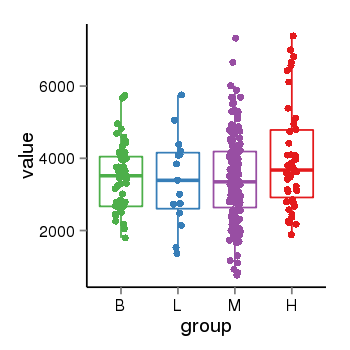 |
| 36611 | tcgaexp | gene=KAZN, entrez=23254, pos=chr1:14925213-15444544(+), B=906, L=624, M=603, H=638 | 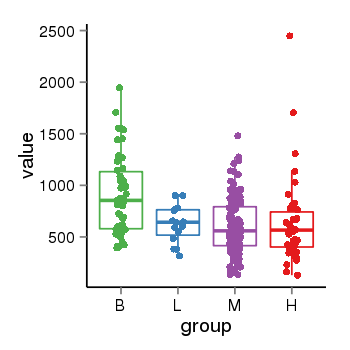 |
| 36611 | tcgaexp | gene=LOC100133331, entrez=100133331, pos=chr1:322037-180753553(-), B=268, L=328, M=271, H=305 | 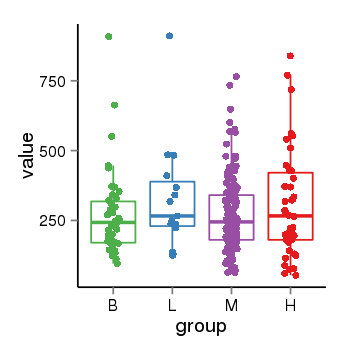 |
| 36611 | pubmed | gene=KAZN, G=7, GP=0, GC=0, GM=0, GPM=0, GCM=0 |  |