DMR 36745 chr1:41948951-41949200 (250bp)
| DMR Stats | |
|---|---|
| Position | chr1:41948951-41949200 (View on UCSC) |
| Width | 250bp |
| Genomic Context | intron |
| Nearest Genes | EDN2 (0bp away) |
| ANODEV p-value | 0.0105659452035 |
| Frequencies | Benign: 0.0 (0.0%), Low: 0.0 (0.0%), High: 7.0 (77.78 %) |
| Frequent? |
Region Count Boxplot

Region Overview

Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 36745 | EDN2 | 735 | uc001cgu.3 | chr1 | 41944446 | 41949874 | - | 250 | 100.0 | 0.23 | 1 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (100), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0) | 0.0 |
| 36745 | EDN2 | 735 | uc001cgv.3 | chr1 | 41944446 | 41949874 | - | 250 | 100.0 | 0.14 | 1 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (100), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0) | 0.0 |
| 36745 | EDN2 | 735 | uc001cgw.3 | chr1 | 41944446 | 41949874 | - | 250 | 100.0 | 0.23 | 1 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0) | 0.0 |
| 36745 | EDN2 | 735 | uc001cgx.3 | chr1 | 41944446 | 41950344 | - | 250 | 100.0 | 0.13 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (100), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0) | 0.0 |
| 36745 | EDN2 | 734 | uc009vwh.3 | chr1 | 41944446 | 41949874 | - | 250 | 100.0 | 0.21 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0) | 0.0 |
| 36745 | EDN2 | 735 | uc009vwi.3 | chr1 | 41944446 | 41949874 | - | 250 | 100.0 | 0.13 | 1 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0) | 0.0 |
| 36745 | EDN2 | 735 | uc009vwj.3 | chr1 | 41944446 | 41949874 | - | 250 | 100.0 | 0.21 | 1 | 0.0 | E1 (0), I1 (100), E2 (0) | 0.0 |
- 7 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 36745 | chr1 | 41947961 | 41949315 | wgEncodeRegDnaseClusteredV2 | 58 | score:1000, srow:33308 |
| 36745 | chr1 | 41948645 | 41949054 | wgEncodeRegTfbsClusteredV3 | GATA2 | score:240, expCount:1, expNums:586, expScores:240, srow:118514 |
| 36745 | chr1 | 41948973 | 41948990 | tfbsConsSites | V$NF1_Q6 | score:878, zScore:1.81, srow:99288 |
| 36745 | chr1 | 41948979 | 41948993 | tfbsConsSites | V$E2F_01 | score:781, zScore:2.14, srow:99289 |
| 36745 | chr1 | 41948981 | 41948992 | tfbsConsSites | V$IK2_01 | score:949, zScore:2, srow:99290 |
| 36745 | chr1 | 41949004 | 41949020 | tfbsConsSites | V$HNF1_C | score:811, zScore:2.01, srow:99291 |
| 36745 | chr1 | 41949044 | 41949059 | tfbsConsSites | V$EVI1_01 | score:752, zScore:1.83, srow:99292 |
| 36745 | chr1 | 41949050 | 41949068 | tfbsConsSites | V$OCT1_01 | score:821, zScore:2.32, srow:99293 |
| 36745 | chr1 | 41949051 | 41949064 | tfbsConsSites | V$CEBPB_01 | score:887, zScore:1.77, srow:99294 |
| 36745 | chr1 | 41949051 | 41949064 | tfbsConsSites | V$CEBP_Q2 | score:955, zScore:3.24, srow:99295 |
| 36745 | chr1 | 41948946 | 41949077 | phastConsElements100way | lod=380 | score:581, srow:181543 |
| 36745 | chr1 | 41949086 | 41949100 | phastConsElements100way | lod=24 | score:308, srow:181544 |
| 36745 | chr1 | 41949109 | 41949132 | phastConsElements100way | lod=105 | score:454, srow:181545 |
| 36745 | chr1 | 41949137 | 41949144 | phastConsElements100way | lod=28 | score:323, srow:181546 |
| 36745 | chr1 | 41949150 | 41949157 | phastConsElements100way | lod=21 | score:295, srow:181547 |
| 36745 | chr1 | 41949184 | 41949189 | phastConsElements100way | lod=17 | score:274, srow:181548 |
- 16 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 36745 | cellmeth_recounts | PrEC: 0, LNCaP: 0, DU145: 0 |  |
| 36745 | cellexp | id=ENSG00000127129, name=EDN2, PrEC=4, str=7, LNCaP=105, str=60, DU145=292, str=204 |  |
| 36745 | tcgaexp | gene=EDN2, entrez=1907, pos=chr1:41944446-41950344(-), B=332, L=185, M=172, H=155 |  |
| 36745 | tcgaexp | gene=LOC100133331, entrez=100133331, pos=chr1:322037-180753553(-), B=268, L=328, M=271, H=305 | 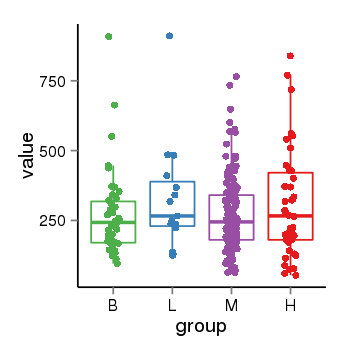 |