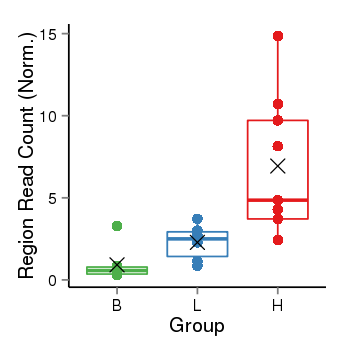
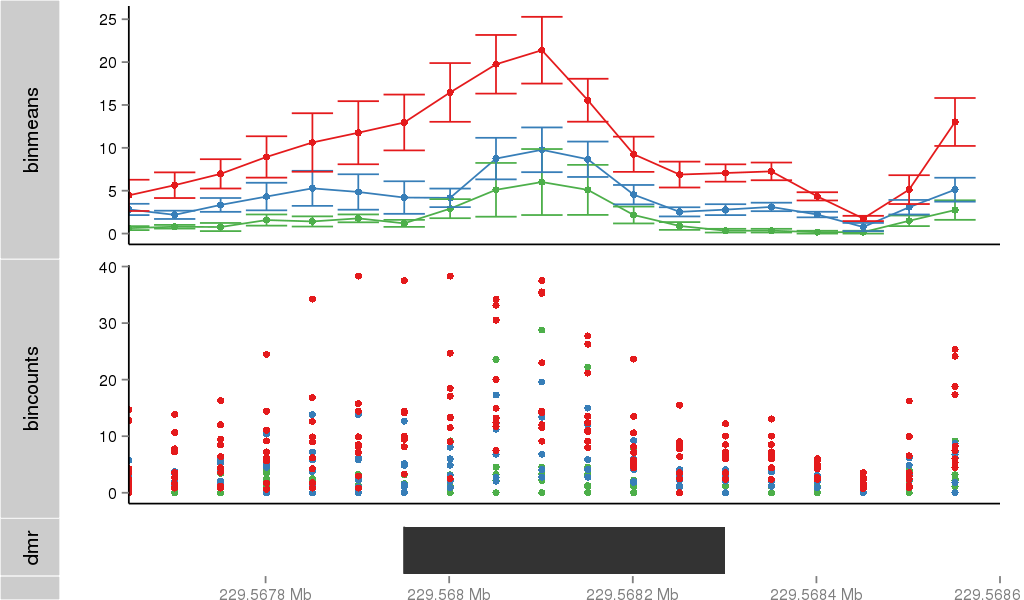
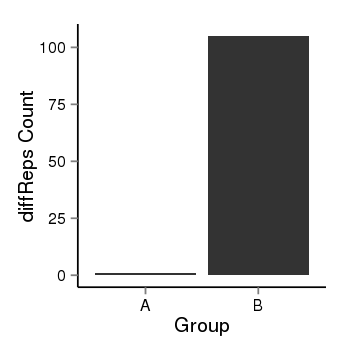
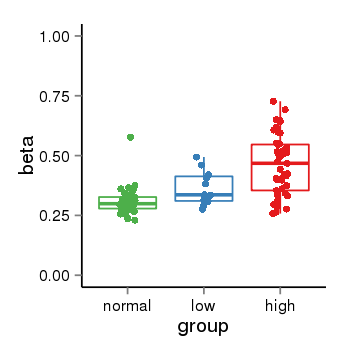
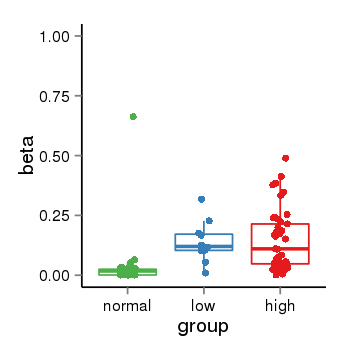
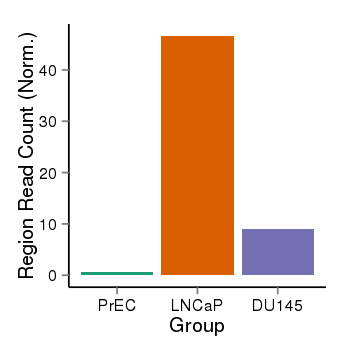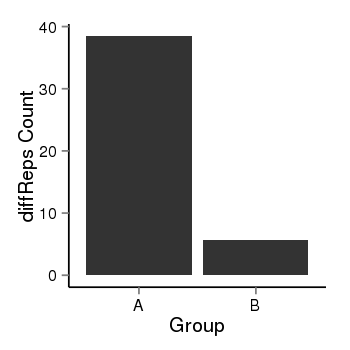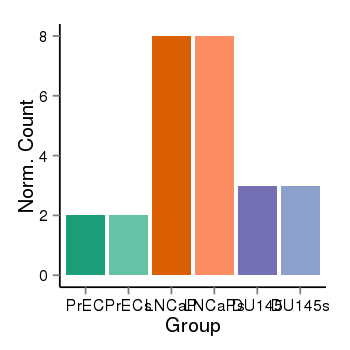| 37019 | chr1 | 229567546 | 229568375 | wgEncodeRegDnaseClusteredV2 | 25 | score:265, srow:110236 |
| 37019 | chr1 | 229566680 | 229568247 | wgEncodeRegTfbsClusteredV3 | EZH2 | score:753, expCount:8, expNums:6,15,19,21,36,38,40,43, expScores:296,253,93,319,615,300,452,538, srow:405386 |
| 37019 | chr1 | 229567884 | 229568921 | wgEncodeRegTfbsClusteredV3 | SUZ12 | score:181, expCount:1, expNums:578, expScores:169, srow:405389 |
| 37019 | chr1 | 229568016 | 229568043 | tfbsConsSites | V$PAX5_01 | score:761, zScore:1.66, srow:476889 |
| 37019 | chr1 | 229568018 | 229568023 | tfbsConsSites | V$AML1_01 | score:1000, zScore:1.78, srow:476890 |
| 37019 | chr1 | 229568074 | 229568093 | tfbsConsSites | V$P53_01 | score:694, zScore:2.04, srow:476891 |
| 37019 | chr1 | 229568088 | 229568108 | tfbsConsSites | V$PAX4_01 | score:850, zScore:2.61, srow:476892 |
| 37019 | chr1 | 229568090 | 229568108 | tfbsConsSites | V$PAX2_01 | score:775, zScore:1.8, srow:476893 |
| 37019 | chr1 | 229568096 | 229568119 | tfbsConsSites | V$HTF_01 | score:765, zScore:1.77, srow:476894 |
| 37019 | chr1 | 229568100 | 229568116 | tfbsConsSites | V$XBP1_01 | score:838, zScore:2.71, srow:476895 |
| 37019 | chr1 | 229568105 | 229568122 | tfbsConsSites | V$CEBP_C | score:858, zScore:2.8, srow:476896 |
| 37019 | chr1 | 229568123 | 229568140 | tfbsConsSites | V$SRF_01 | score:787, zScore:2.81, srow:476897 |
| 37019 | chr1 | 229568125 | 229568141 | tfbsConsSites | V$YY1_01 | score:859, zScore:1.67, srow:476898 |
| 37019 | chr1 | 229568129 | 229568152 | tfbsConsSites | V$HTF_01 | score:761, zScore:1.69, srow:476899 |
| 37019 | chr1 | 229568134 | 229568147 | tfbsConsSites | V$MYCMAX_01 | score:802, zScore:1.8, srow:476900 |
| 37019 | chr1 | 229568041 | 229568041 | cosmic | COSM905240 | srow:116199 |
| 37019 | chr1 | 229568069 | 229568069 | cosmic | COSM905241 | srow:116200 |
| 37019 | chr1 | 229568092 | 229568092 | cosmic | COSM414578 | srow:116201 |
| 37019 | chr1 | 229568141 | 229568141 | cosmic | COSM905242 | srow:116202 |
| 37019 | chr1 | 229568145 | 229568145 | cosmic | COSM533316 | srow:116203 |
| 37019 | chr1 | 229568165 | 229568165 | cosmic | COSM1601863 | srow:116204 |
| 37019 | chr1 | 229568170 | 229568170 | cosmic | COSM1501225 | srow:116205 |
| 37019 | chr1 | 229568014 | 229568044 | phastConsElements100way | lod=383 | score:582, srow:815210 |
| 37019 | chr1 | 229568046 | 229568068 | phastConsElements100way | lod=345 | score:572, srow:815211 |
| 37019 | chr1 | 229568070 | 229568083 | phastConsElements100way | lod=171 | score:502, srow:815212 |