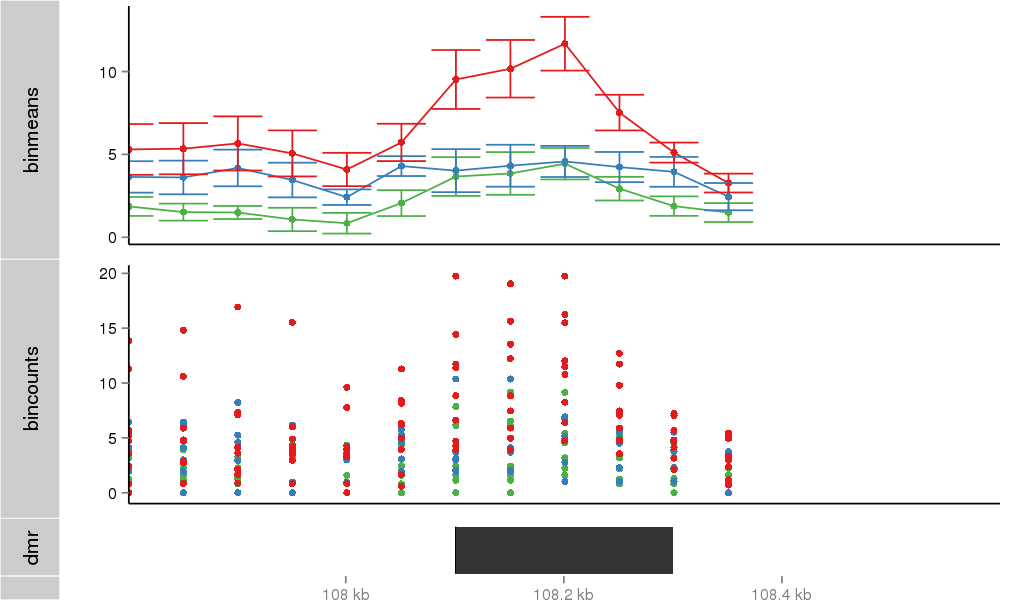
DMR 37723 chr4:108101-108300 (200bp)
| DMR Stats | |
|---|---|
| Position | chr4:108101-108300 (View on UCSC) |
| Width | 200bp |
| Genomic Context | intron |
| Nearest Genes | ZNF718, ZNF595 (0bp away) |
| ANODEV p-value | 0.00228068274232 |
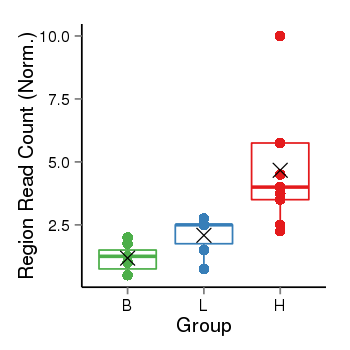
| Frequencies | Benign: 0.0 (0.0%), Low: 0.0 (0.0%), High: 7.0 (77.78 %) |
| Frequent? |
Region Count Boxplot
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 37723 | ZNF718 | 21252 | uc003fzt.4 | chr4 | 53227 | 156490 | + | 200 | 100.0 | 0.07 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (100), E4 (0) | 0.0 |
| 37723 | ZNF595 | 21252 | uc003fzu.1 | chr4 | 53227 | 196092 | + | 200 | 100.0 | 0.09 | 1 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (100), E4 (0), I4 (0), E5 (0) | 0.0 |
- 2 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 37723 | chr4 | 107826 | 108455 | wgEncodeRegDnaseClusteredV2 | 16 | score:367, srow:864228 |
| 37723 | chr4 | 106943 | 108322 | wgEncodeRegTfbsClusteredV3 | EZH2 | score:863, expCount:6, expNums:6,15,19,21,36,38, expScores:510,438,145,404,317,196, srow:2988382 |
| 37723 | chr4 | 107899 | 109898 | cpgShore | — | values(x.gr)[overs$srow, ]:34970 |
- 3 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 37723 | cellmeth_recounts | PrEC: 0, LNCaP: 0, DU145: 4 | 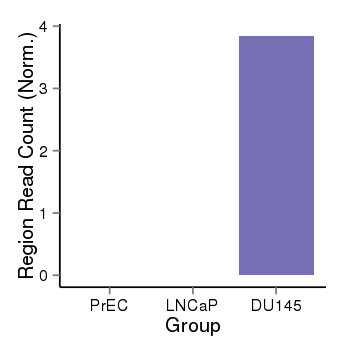 |
| 37723 | tcgaexp | gene=YTHDC1, entrez=91746, pos=chr4:6027-69215824(-), B=4512, L=3976, M=4109, H=4458 | 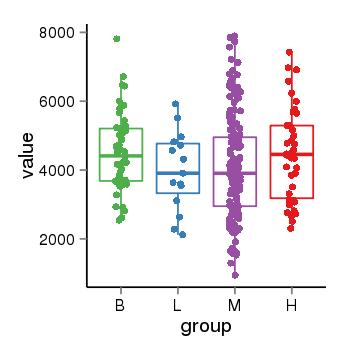 |
| 37723 | tcgaexp | gene=ZNF595, entrez=152687, pos=chr4:53227-196092(+), B=502, L=637, M=656, H=769 | 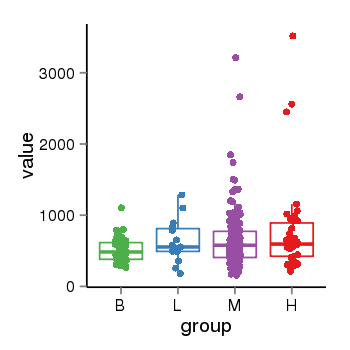 |
| 37723 | tcgaexp | gene=ZNF718, entrez=255403, pos=chr4:53227-156490(+), B=244, L=237, M=227, H=291 | 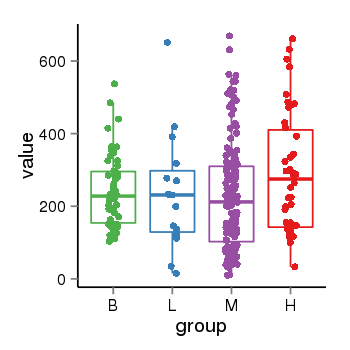 |
| 37723 | tcgaexp | gene=DUX4L5, entrez=653545, pos=chr4:72836-191010142(+), B=0, L=0, M=0, H=0 |  |