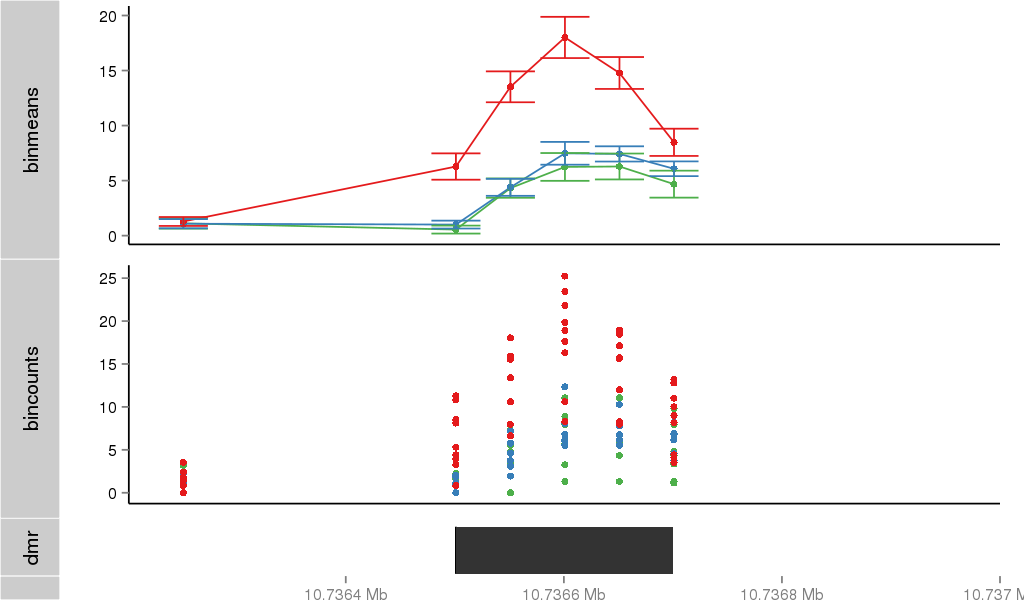
DMR 37988 chr5:10736501-10736700 (200bp)
| DMR Stats | |
|---|---|
| Position | chr5:10736501-10736700 (View on UCSC) |
| Width | 200bp |
| Genomic Context | intron |
| Nearest Genes | DAP (0bp away) |
| ANODEV p-value | 1.21800254835e-06 |
| Frequencies | Benign: 0.0 (0.0%), Low: 0.0 (0.0%), High: 8.0 (88.89 %) |
| Frequent? |
Region Count Boxplot

Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 37988 | DAP | 22451 | uc003jez.4 | chr5 | 10679342 | 10761387 | - | 200 | 100.0 | 0.03 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (100), E3 (0), I3 (0), E4 (0) | 0.0 |
| 37988 | DAP | 22451 | uc011cmw.2 | chr5 | 10679342 | 10761387 | - | 200 | 100.0 | 0.02 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (100), E3 (0) | 0.0 |
- 2 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 37988 | chr5 | 10736426 | 10736875 | wgEncodeRegDnaseClusteredV2 | 7 | score:236, srow:930900 |
| 37988 | chr5 | 10736101 | 10736578 | wgEncodeRegTfbsClusteredV3 | CTCF | score:413, expCount:17, expNums:2,5,24,213,290,493,601,603,604,605,606,621,651,653,654,656,679, expScores:132,115,162,190,192,393,125,399,211,413,235,290,101,148,151,106,101, srow:3179870 |
| 37988 | chr5 | 10736295 | 10736818 | wgEncodeRegTfbsClusteredV3 | HDAC1 | score:174, expCount:1, expNums:26, expScores:174, srow:3179873 |
- 3 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 37988 | cellmeth_recounts | PrEC: 1, LNCaP: 0, DU145: 0 | 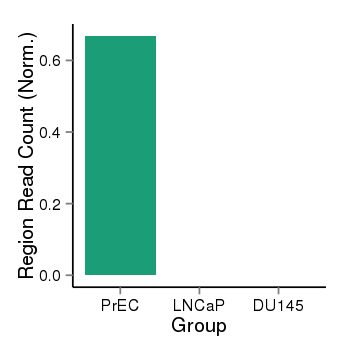 |
| 37988 | cellexp | id=ENSG00000112977, name=DAP, PrEC=7211, str=4678, LNCaP=9193, str=11217, DU145=10177, str=11814 | 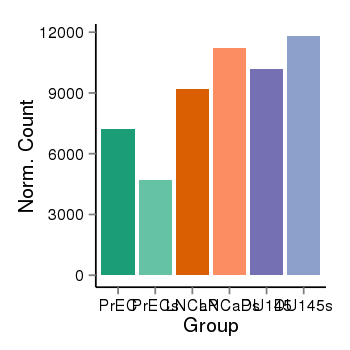 |
| 37988 | tcgaexp | gene=DAP, entrez=1611, pos=chr5:10679342-10761387(-), B=13420, L=12641, M=13038, H=12853 | 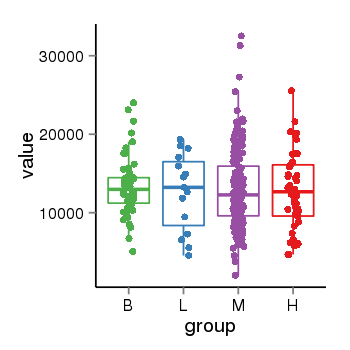 |
| 37988 | pubmed | gene=DAP, G=3603, GP=21, GC=426, GM=114, GPM=0, GCM=103 | 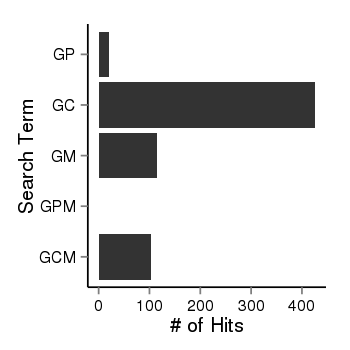 |