DMR 40306 chr12:112282951-112283150 (200bp)
| DMR Stats | |
|---|---|
| Position | chr12:112282951-112283150 (View on UCSC) |
| Width | 200bp |
| Genomic Context | intron |
| Nearest Genes | MAPKAPK5 (0bp away) |
| ANODEV p-value | 1.76290620231e-05 |
| Frequencies | Benign: 0.0 (0.0%), Low: 0.0 (0.0%), High: 6.0 (66.67 %) |
| Frequent? |
Region Count Boxplot

Region Overview

Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 40306 | MAPKAPK5 | 7039 | uc001tsz.4 | chr12 | 112280032 | 112331228 | + | 200 | 100.0 | 0.08 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0), I8 (0), E9 (0), I9 (0), E10 (0), I10 (0), E11 (0), I11 (0), E12 (0), I12 (0), E13 (0), I13 (0), E14 (0) | 0.0 |
| 40306 | MAPKAPK5 | 7039 | uc001tta.4 | chr12 | 112280032 | 112331228 | + | 200 | 100.0 | 0.05 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0), I8 (0), E9 (0), I9 (0), E10 (0), I10 (0), E11 (0), I11 (0), E12 (0), I12 (0), E13 (0), I13 (0), E14 (0) | 0.0 |
- 2 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 40306 | chr12 | 112282646 | 112282955 | wgEncodeRegDnaseClusteredV2 | 8 | score:262, srow:296117 |
| 40306 | chr12 | 112282715 | 112284714 | cpgShelf | — | values(x.gr)[overs$srow, ]:9270 |
- 2 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 40306 | cellmeth_recounts | PrEC: 6, LNCaP: 5, DU145: 10 |  |
| 40306 | cellexp | id=ENSG00000089022, name=MAPKAPK5, PrEC=1929, str=1472, LNCaP=1569, str=2114, DU145=1587, str=2133 |  |
| 40306 | tcgaexp | gene=ZNF26, entrez=7574, pos=chr12:93510-133589154(+), B=419, L=392, M=412, H=512 | 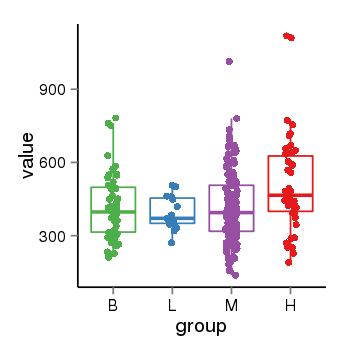 |
| 40306 | tcgaexp | gene=ZNF84, entrez=7637, pos=chr12:42780-133639885(+), B=1626, L=1649, M=1716, H=2045 | 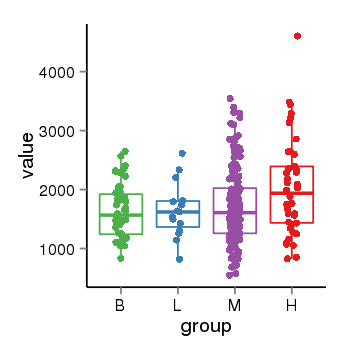 |
| 40306 | tcgaexp | gene=MAPKAPK5, entrez=8550, pos=chr12:112280032-112331228(+), B=1321, L=1445, M=1450, H=1594 |  |
| 40306 | pubmed | gene=MAPKAPK5, G=62, GP=0, GC=13, GM=0, GPM=0, GCM=0 |  |