DMR 40819 chr15:75945701-75945900 (200bp)
| DMR Stats | |
|---|---|
| Position | chr15:75945701-75945900 (View on UCSC) |
| Width | 200bp |
| Genomic Context | intron |
| Nearest Genes | SNX33 (0bp away) |
| ANODEV p-value | 5.61849412307e-08 |
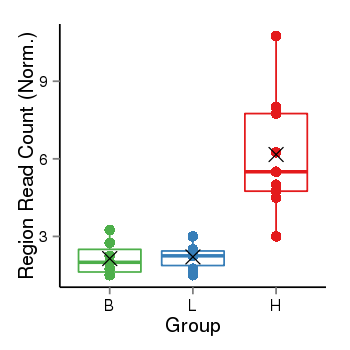
| Frequencies | Benign: 1.0 (14.29%), Low: 0.0 (0.0%), High: 8.0 (88.89 %) |
| Frequent? |
Region Count Boxplot
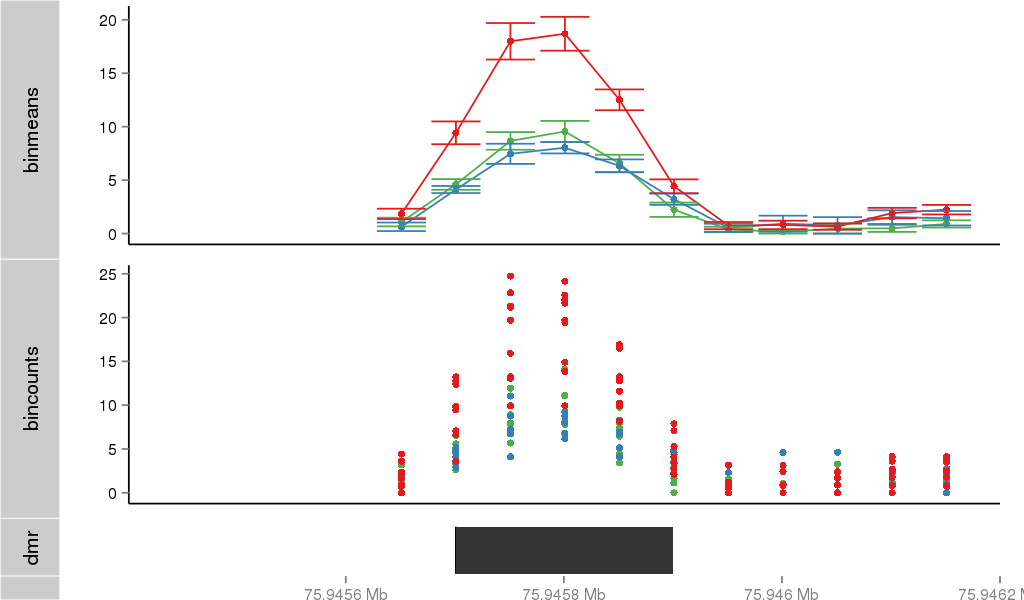
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 40819 | SNX33 | 10016 | uc002bau.3 | chr15 | 75941348 | 75950968 | + | 200 | 100.0 | 0.59 | 0 | 0.0 | E1 (0), I1 (100), E2 (0) | 0.0 |
| 40819 | SNX33 | 10016 | uc002bav.3 | chr15 | 75942097 | 75950968 | + | 200 | 100.0 | 0.3 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (100), E3 (0) | 0.0 |
- 2 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 40819 | chr15 | 75945646 | 75946070 | wgEncodeRegDnaseClusteredV2 | 8 | score:385, srow:412139 |
| 40819 | chr15 | 75945795 | 75946091 | wgEncodeRegTfbsClusteredV3 | ZNF263 | score:420, expCount:1, expNums:368, expScores:420, srow:1396541 |
| 40819 | chr15 | 75945725 | 75945727 | phastConsElements100way | lod=13 | score:247, srow:3117056 |
| 40819 | chr15 | 75945862 | 75945867 | phastConsElements100way | lod=13 | score:247, srow:3117057 |
- 4 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 40819 | cellmeth_recounts | PrEC: 23, LNCaP: 4, DU145: 1 | 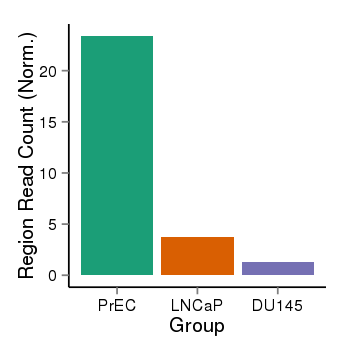 |
| 40819 | cellmeth_diffreps_PrECvsLNCaP | A=35, B=3, Length=380, Event=Down, log2FC=-3.54, padj=1.14208112046554e-06 | 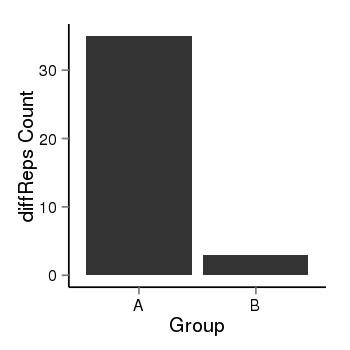 |
| 40819 | cellmeth_diffreps_PrECvsDU145 | A=35, B=1, Length=420, Event=Down, log2FC=-5.13, padj=1.81683967010991e-08 | 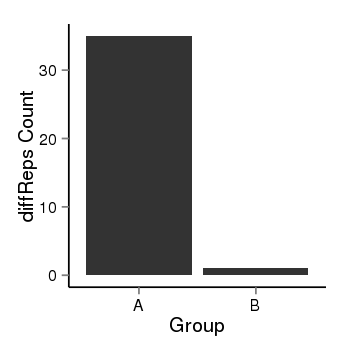 |
| 40819 | cellexp | id=ENSG00000173548, name=SNX33, PrEC=5032, str=8680, LNCaP=1842, str=958, DU145=3096, str=1675 | 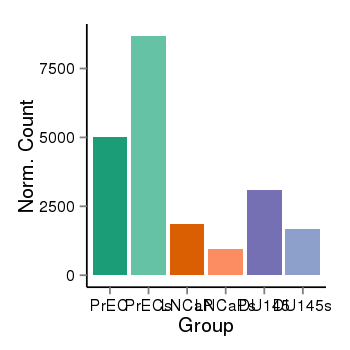 |
| 40819 | tcgaexp | gene=SNX33, entrez=257364, pos=chr15:75941348-75950968(+), B=2953, L=2832, M=2782, H=2350 | 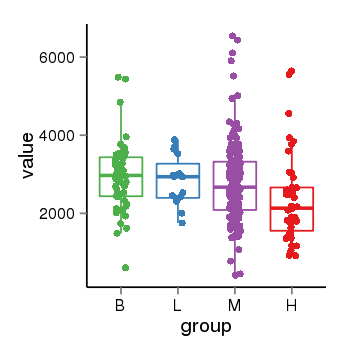 |
| 40819 | pubmed | gene=SNX33, G=9, GP=0, GC=1, GM=0, GPM=0, GCM=0 | 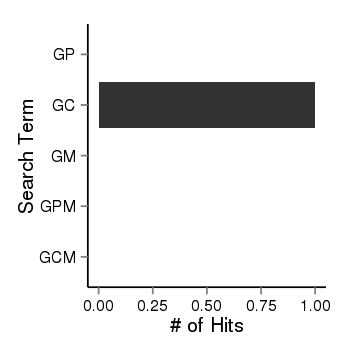 |