| 41087 | cellmeth_recounts | PrEC: 0, LNCaP: 0, DU145: 1 |  |
| 41087 | cellexp | id=ENSG00000171861, name=RNMTL1, PrEC=965, str=485, LNCaP=1634, str=1944, DU145=1161, str=1377 |  |
| 41087 | cellexp | id=ENSG00000262434, name=RP11-676J12.8, PrEC=8, str=28, LNCaP=4, str=5, DU145=29, str=37 |  |
| 41087 | tcgaexp | gene=NSF, entrez=4905, pos=chr17:266212-44834828(+), B=3355, L=5678, M=5410, H=5700 | 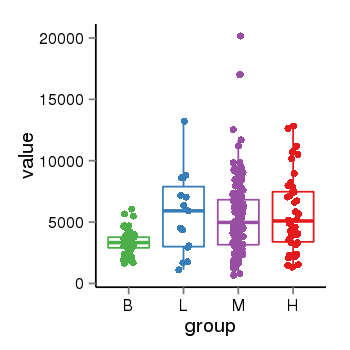 |
| 41087 | tcgaexp | gene=PLEKHM1, entrez=9842, pos=chr17:128328-43568146(-), B=1993, L=1631, M=1589, H=1418 | 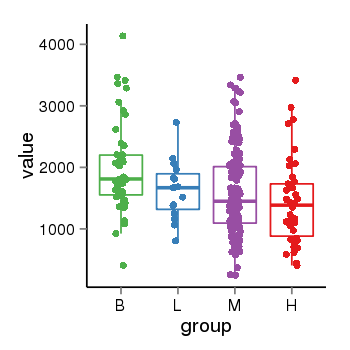 |
| 41087 | tcgaexp | gene=LRRC37A, entrez=9884, pos=chr17:415627-44415160(+), B=924, L=896, M=971, H=940 | 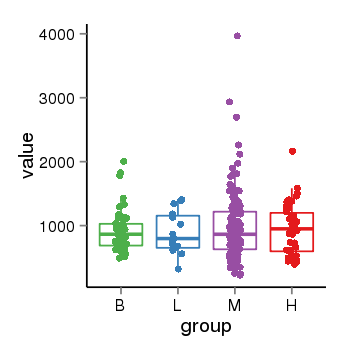 |
| 41087 | tcgaexp | gene=ARL17A, entrez=51326, pos=chr17:195721-44657088(-), B=200, L=195, M=210, H=230 | 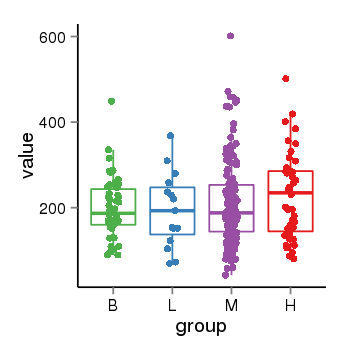 |
| 41087 | tcgaexp | gene=LRRC37A4P, entrez=55073, pos=chr17:197706-43597889(-), B=1015, L=1512, M=1396, H=1450 | 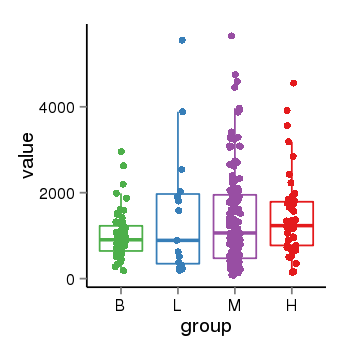 |
| 41087 | tcgaexp | gene=RNMTL1, entrez=55178, pos=chr17:685513-695741(+), B=817, L=1068, M=986, H=777 |  |
| 41087 | tcgaexp | gene=ARHGAP27, entrez=201176, pos=chr17:86404-43511112(-), B=1570, L=1264, M=1273, H=1486 | 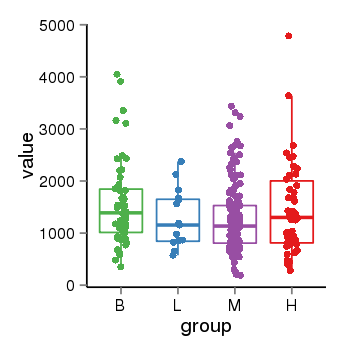 |
| 41087 | tcgaexp | gene=KANSL1, entrez=284058, pos=chr17:563498-44302740(-), B=3141, L=2594, M=2797, H=3017 | 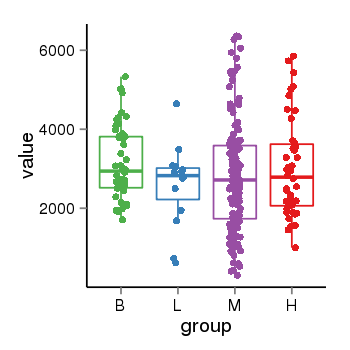 |
| 41087 | tcgaexp | gene=LRRC37A2, entrez=474170, pos=chr17:197706-44633014(+), B=351, L=310, M=361, H=377 | 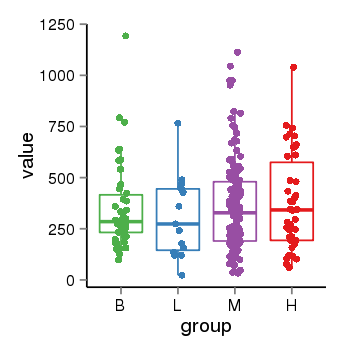 |













