DMR 4205 chr7:139102501-139102750 (250bp)
| DMR Stats | |
|---|---|
| Position | chr7:139102501-139102750 (View on UCSC) |
| Width | 250bp |
| Genomic Context | 3' end |
| Nearest Genes | LUC7L2, C7orf55-LUC7L2, LOC100129148 (0bp away) |
| ANODEV p-value | 0.0103738848817 |
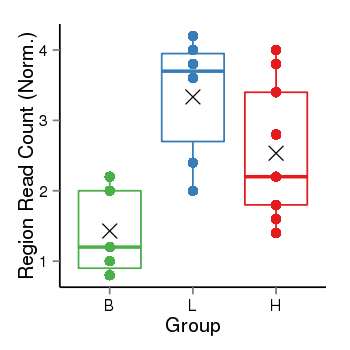
| Frequencies | Benign: 0.0 (0.0%), Low: 4.0 (66.67%), High: 3.0 (33.33 %) |
| Frequent? |
Region Count Boxplot
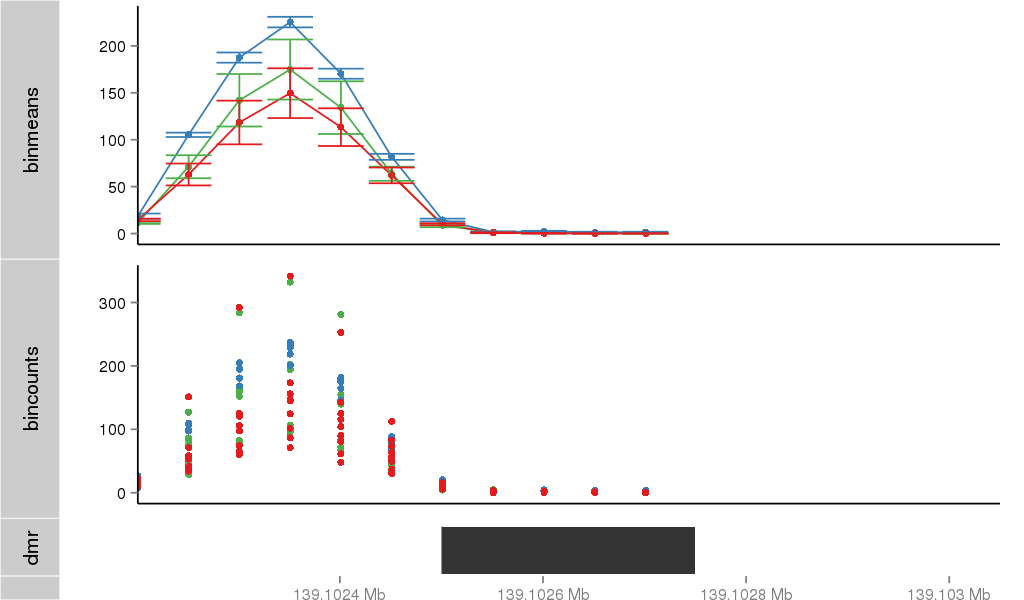
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4205 | LUC7L2 | 27784 | uc003vux.4 | chr7 | 139044592 | 139108203 | + | 250 | 100.0 | 0.03 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0), I8 (0), E9 (0), I9 (100), E10 (0) | 0.0 |
| 4205 | LUC7L2 | 27784 | uc003vuy.4 | chr7 | 139044592 | 139108203 | + | 250 | 100.0 | 0.03 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0), I8 (0), E9 (0), I9 (0), E10 (0), I10 (100), E11 (0) | 0.0 |
| 4205 | LUC7L2 | 27784 | uc003vva.3 | chr7 | 139059419 | 139108203 | + | 250 | 100.0 | 0.03 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0), I8 (100), E9 (0) | 0.0 |
| 4205 | LUC7L2 | 27784 | uc011kqs.2 | chr7 | 139025105 | 139108203 | + | 250 | 100.0 | 0.03 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0), I8 (0), E9 (0), I9 (0), E10 (0), I10 (100), E11 (0) | 0.0 |
| 4205 | C7orf55-LUC7L2 | 27784 | uc011kqt.3 | chr7 | 139025878 | 139108203 | + | 250 | 100.0 | 0.03 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0), I8 (0), E9 (0), I9 (0), E10 (0), I10 (100), E11 (0) | 0.0 |
| 4205 | LOC100129148 | 27786 | uc022ams.1 | chr7 | 139102209 | 139112272 | - | 250 | 100.0 | 0.03 | 1 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (100), E3 (0) | 100.0 |
- 6 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 4205 | chr7 | 139102202 | 139102605 | wgEncodeRegTfbsClusteredV3 | POLR2A | score:369, expCount:2, expNums:228,229, expScores:215,369, srow:3888859 |
| 4205 | chr7 | 139102561 | 139102572 | tfbsConsSites | V$CDP_01 | score:842, zScore:2.96, srow:4996876 |
| 4205 | chr7 | 139102564 | 139102575 | tfbsConsSites | V$FOXD3_01 | score:926, zScore:2.43, srow:4996877 |
| 4205 | chr7 | 139102697 | 139102710 | tfbsConsSites | V$COUP_01 | score:849, zScore:1.77, srow:4996878 |
| 4205 | chr7 | 139102521 | 139102545 | phastConsElements100way | lod=38 | score:354, srow:8642019 |
| 4205 | chr7 | 139102570 | 139102575 | phastConsElements100way | lod=27 | score:320, srow:8642020 |
| 4205 | chr7 | 139102590 | 139102591 | phastConsElements100way | lod=13 | score:247, srow:8642021 |
| 4205 | chr7 | 139102598 | 139102614 | phastConsElements100way | lod=48 | score:377, srow:8642022 |
| 4205 | chr7 | 139102679 | 139102691 | phastConsElements100way | lod=15 | score:262, srow:8642023 |
| 4205 | chr7 | 139102703 | 139102706 | phastConsElements100way | lod=18 | score:280, srow:8642024 |
| 4205 | chr7 | 139102712 | 139102726 | phastConsElements100way | lod=13 | score:247, srow:8642025 |
- 11 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 4205 | cellmeth_recounts | PrEC: 5, LNCaP: 8, DU145: 19 | 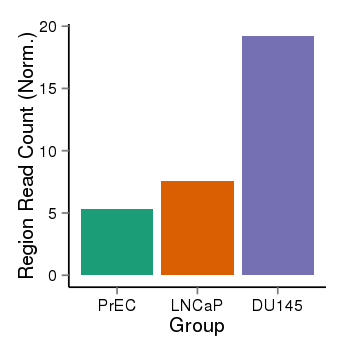 |
| 4205 | cellmeth_diffreps_PrECvsDU145 | A=92, B=27, Length=400, Event=Down, log2FC=-1.77, padj=8.93530311565591e-08 | 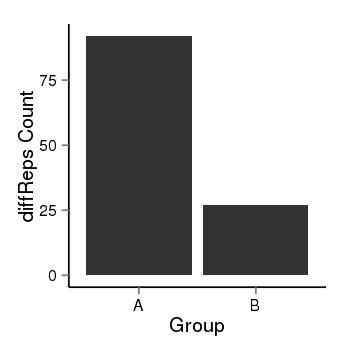 |
| 4205 | cellmeth_diffreps_PrECvsDU145 | A=0, B=11, Length=200, Event=Up, log2FC=Inf, padj=0.00313959514768542 | 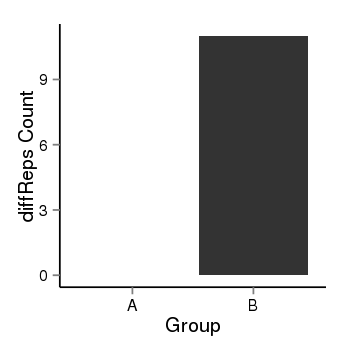 |
| 4205 | cellmeth_diffreps_LNCaPvsDU145 | A=52.38, B=21.51, Length=240, Event=Down, log2FC=-1.28, padj=0.0229250110230626 | 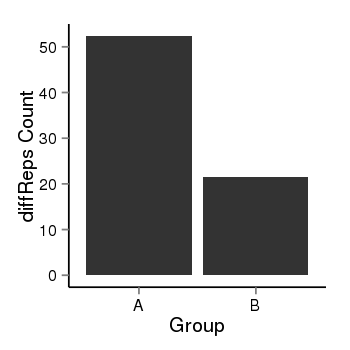 |
| 4205 | cellexp | id=ENSG00000146963, name=C7orf55-LUC7L2, PrEC=1811, str=1895, LNCaP=2362, str=2222, DU145=1636, str=1882 | 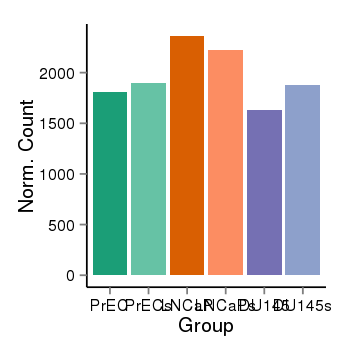 |
| 4205 | cellexp | id=ENSG00000269955, name=LUC7L2, PrEC=0, str=0, LNCaP=0, str=0, DU145=0, str=0 |  |
| 4205 | tcgaexp | gene=LUC7L2, entrez=51631, pos=chr7:139025105-139108203(+), B=5257, L=5202, M=5374, H=6547 | 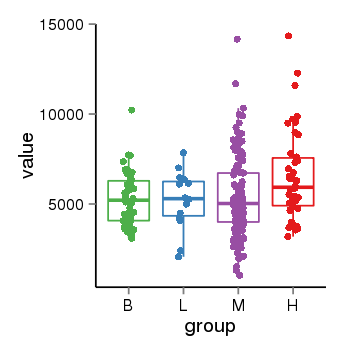 |