DMR 456 chr1:148943351-148943500 (150bp)
| DMR Stats | |
|---|---|
| Position | chr1:148943351-148943500 (View on UCSC) |
| Width | 150bp |
| Genomic Context | intron |
| Nearest Genes | LOC645166 (0bp away) |
| ANODEV p-value | 0.00295465297512 |
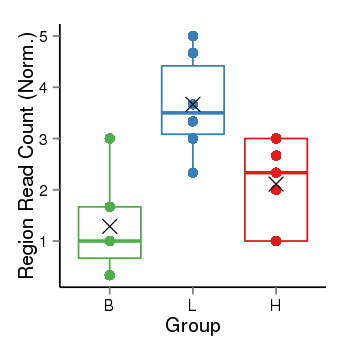
| Frequencies | Benign: 0.0 (0.0%), Low: 4.0 (66.67%), High: 0.0 (0.0 %) |
| Frequent? |
Region Count Boxplot
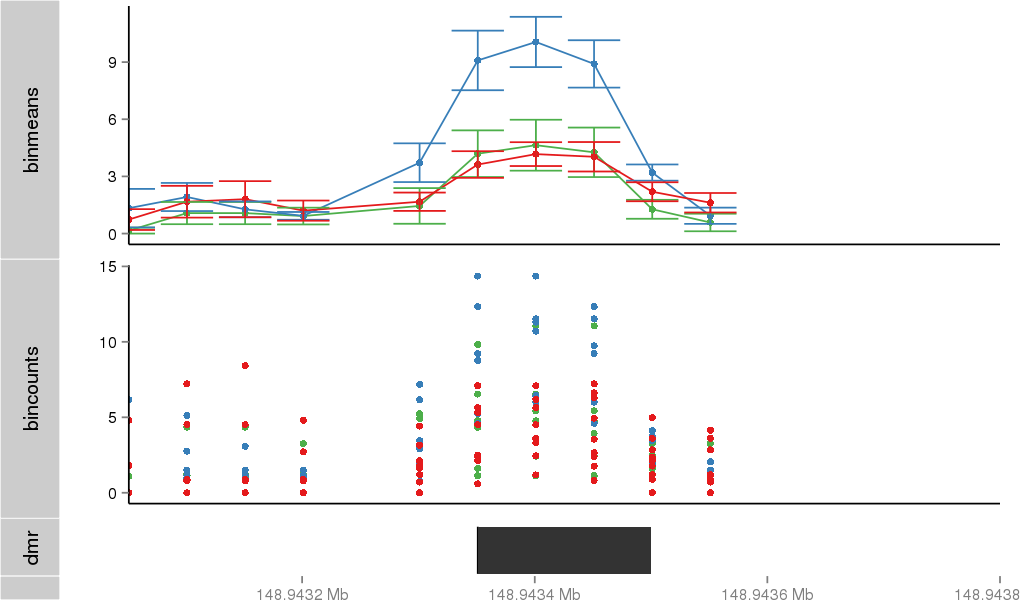
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 456 | LOC645166 | 1567 | uc009wkw.1 | chr1 | 148930405 | 148953054 | + | 150 | 100.0 | 0.41 | 1 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (100), E4 (0) | 0.0 |
| 456 | LOC645166 | 1567 | uc010pbc.1 | chr1 | 148928286 | 148951595 | + | 150 | 100.0 | 0.41 | 1 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (100), E4 (0) | 0.0 |
| 456 | LOC645166 | 1567 | uc010pbd.1 | chr1 | 148928286 | 148951595 | + | 150 | 100.0 | 1.24 | 1 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (100), E3 (0) | 0.0 |
- 3 geness
Feature Overlaps
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 456 | cellmeth_recounts | PrEC: 2, LNCaP: 1, DU145: 20 | 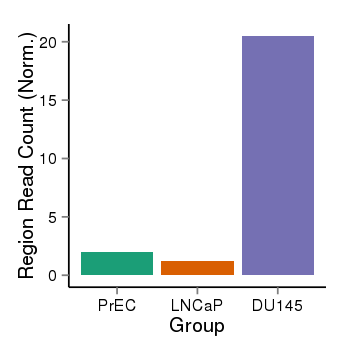 |
| 456 | cellmeth_diffreps_LNCaPvsDU145 | A=0, B=14.97, Length=240, Event=Up, log2FC=Inf, padj=0.000816028517810598 | 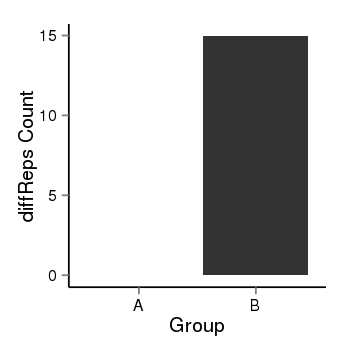 |
| 456 | cellexp | id=ENSG00000232527, name=RP11-14N7.2, PrEC=185, str=291, LNCaP=731, str=524, DU145=591, str=417 | 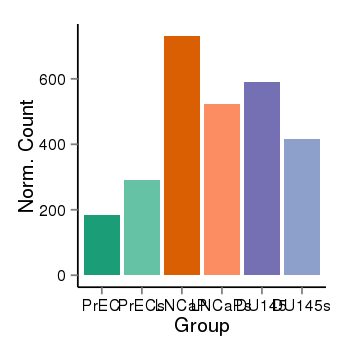 |
| 456 | tcgaexp | gene=LOC645166, entrez=645166, pos=chr1:148928286-148953054(+), B=208, L=248, M=214, H=217 | 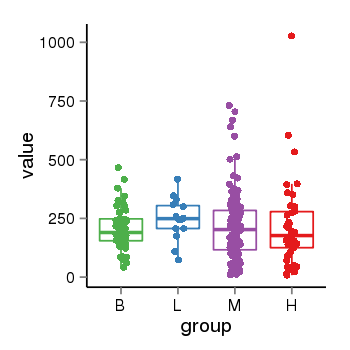 |
| 456 | tcgaexp | gene=PPIAL4B, entrez=653505, pos=chr1:144363462-149553787(-), B=0, L=0, M=0, H=0 | 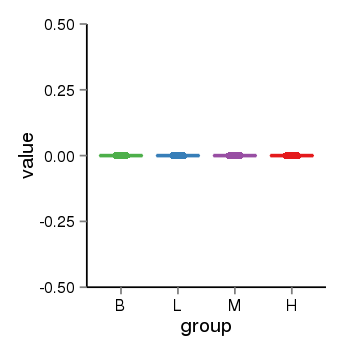 |
| 456 | tcgaexp | gene=LINC00623, entrez=728855, pos=chr1:144300512-149616786(-), B=727, L=966, M=1062, H=1250 | 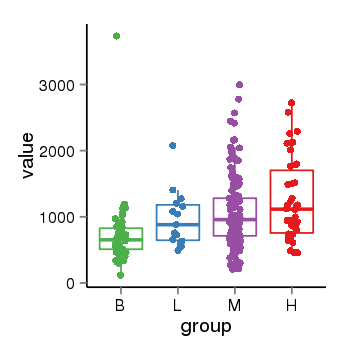 |
| 456 | tcgaexp | gene=LOC100133331, entrez=100133331, pos=chr1:322037-180753553(-), B=268, L=328, M=271, H=305 | 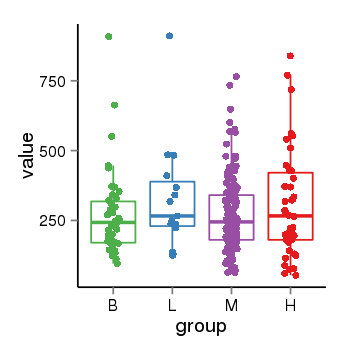 |