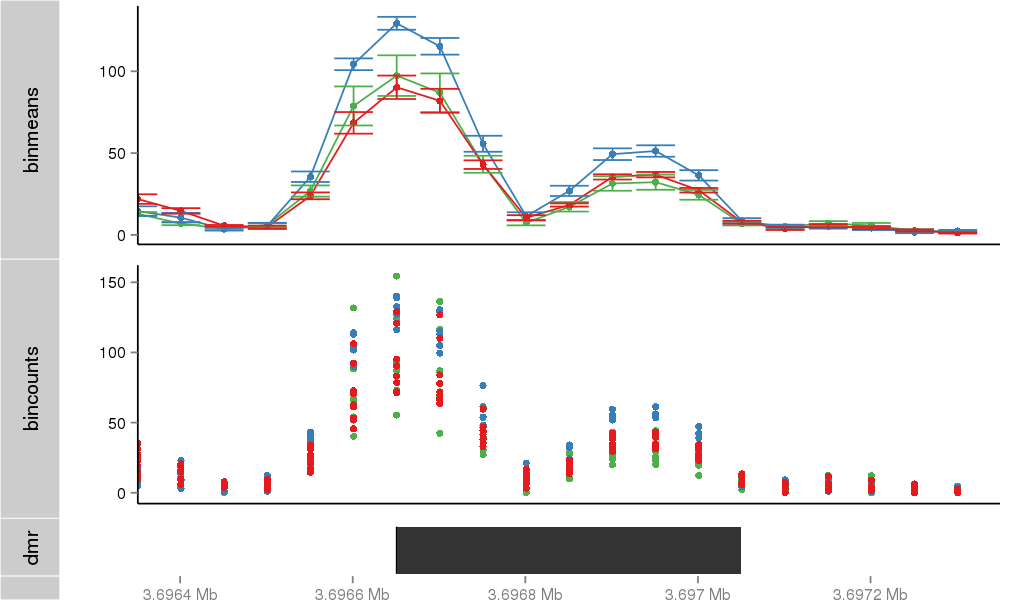
DMR 46 chr1:3696651-3697050 (400bp)
| DMR Stats | |
|---|---|
| Position | chr1:3696651-3697050 (View on UCSC) |
| Width | 400bp |
| Genomic Context | 3' end |
| Nearest Genes | LRRC47 (0bp away) |
| ANODEV p-value | 0.00418422147223 |
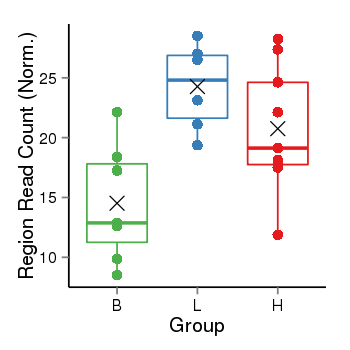
| Frequencies | Benign: 7.0 (100.0%), Low: 6.0 (100.0%), High: 9.0 (100.0 %) |
| Frequent? |
Region Count Boxplot
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 46 | LRRC47 | 128 | uc001akx.1 | chr1 | 3696784 | 3713068 | - | 267 | 66.75 | 1.22 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (66.75) | 100.0 |
- 1 genes
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 46 | chr1 | 3696266 | 3696770 | wgEncodeRegDnaseClusteredV2 | 5 | score:297, srow:2980 |
| 46 | chr1 | 3693507 | 3697419 | wgEncodeRegTfbsClusteredV3 | POLR2A | score:804, expCount:11, expNums:105,191,192,445,446,522,523,526,583,616,622, expScores:541,419,216,102,198,194,187,416,390,353,213, srow:10627 |
- 2 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 46 | cellmeth_recounts | PrEC: 290, LNCaP: 33, DU145: 22 | 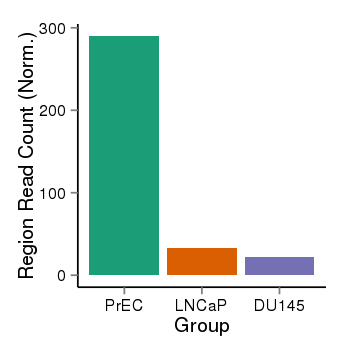 |
| 46 | cellmeth_diffreps_PrECvsLNCaP | A=440, B=26, Length=740, Event=Down, log2FC=-4.08, padj=0 | 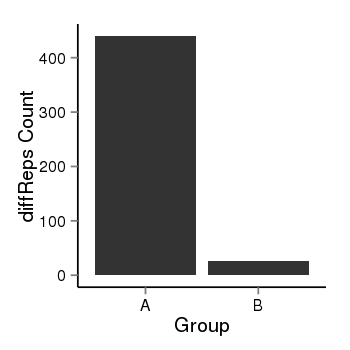 |
| 46 | cellmeth_diffreps_PrECvsDU145 | A=554, B=27, Length=1340, Event=Down, log2FC=-4.36, padj=0 | 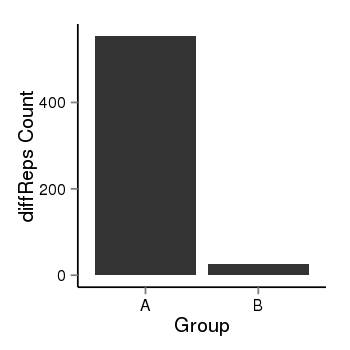 |
| 46 | cellexp | id=ENSG00000130764, name=LRRC47, PrEC=4045, str=2998, LNCaP=4454, str=5124, DU145=1750, str=1898 | 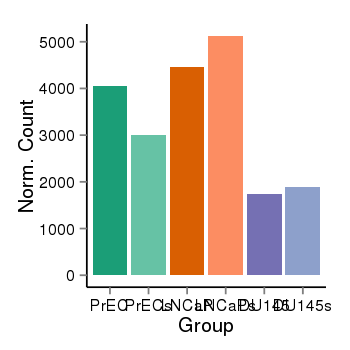 |
| 46 | tcgameth | site:cg03256904, B=0.76, L=0.75, H=0.74 | 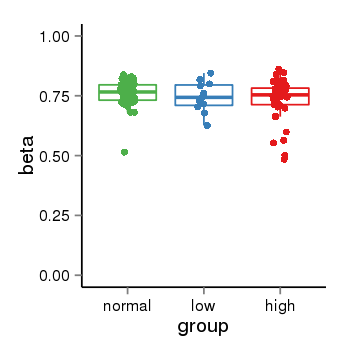 |
| 46 | tcgameth | site:cg25351920, B=0.89, L=0.89, H=0.87 | 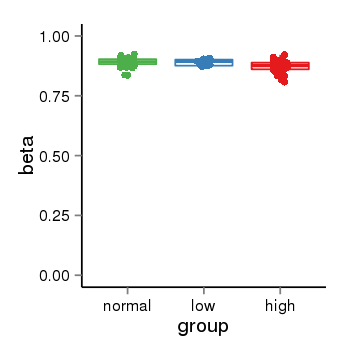 |
| 46 | tcgaexp | gene=SRSF10, entrez=10772, pos=chr1:36273-24306953(-), B=3487, L=3388, M=3432, H=4019 | 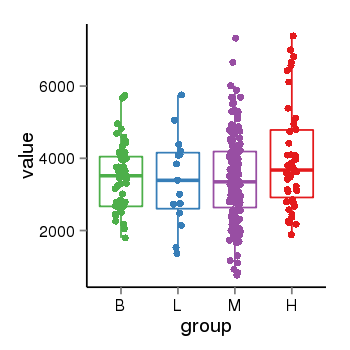 |
| 46 | tcgaexp | gene=LRRC47, entrez=57470, pos=chr1:3696784-3713068(-), B=3945, L=4406, M=3958, H=3852 | 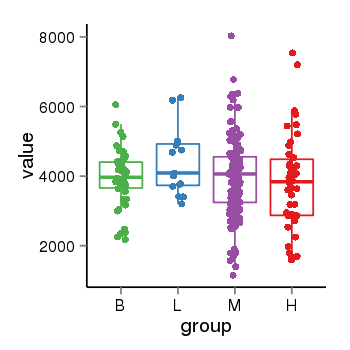 |
| 46 | tcgaexp | gene=LOC100133331, entrez=100133331, pos=chr1:322037-180753553(-), B=268, L=328, M=271, H=305 | 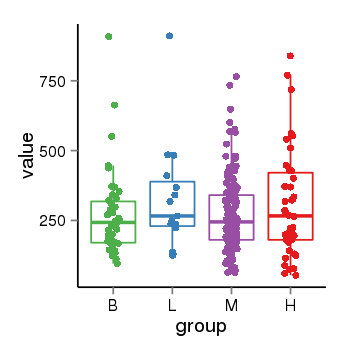 |