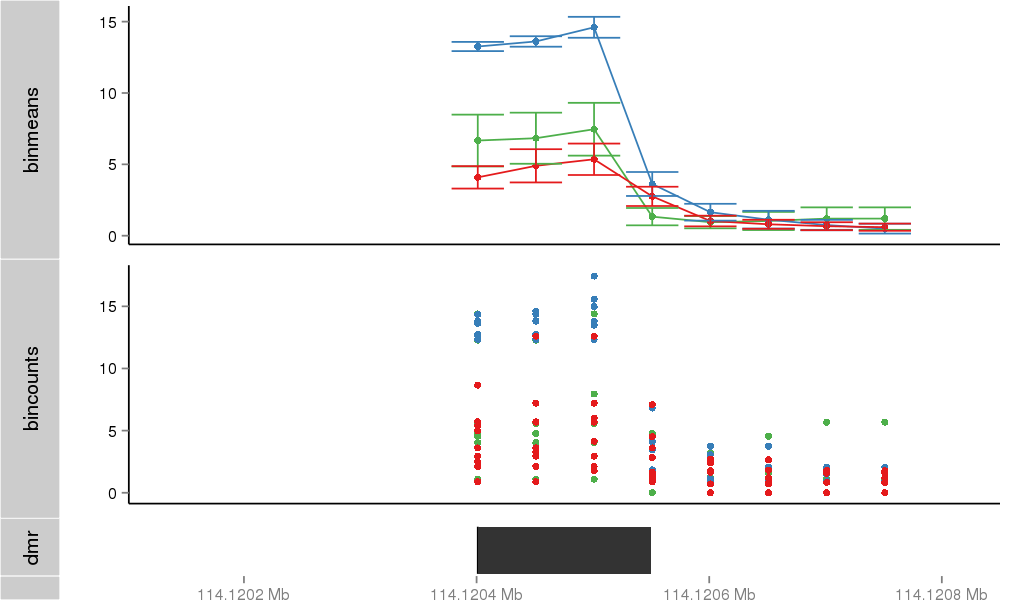
DMR 6092 chr11:114120401-114120550 (150bp)
| DMR Stats | |
|---|---|
| Position | chr11:114120401-114120550 (View on UCSC) |
| Width | 150bp |
| Genomic Context | 3' end |
| Nearest Genes | ZBTB16 (0bp away) |
| ANODEV p-value | 0.000292847047071 |
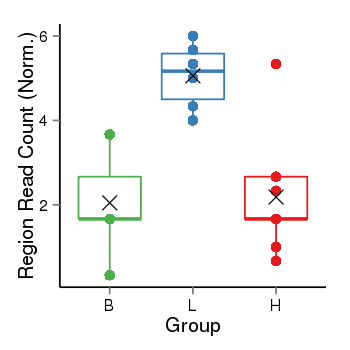
| Frequencies | Benign: 1.0 (14.29%), Low: 6.0 (100.0%), High: 1.0 (11.11 %) |
| Frequent? |
Region Count Boxplot
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6092 | ZBTB16 | 5625 | uc001pop.3 | chr11 | 113930431 | 114121397 | + | 150 | 100.0 | 0.11 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (100), E7 (0) | 100.0 |
| 6092 | ZBTB16 | 5625 | uc001poq.3 | chr11 | 113931288 | 114121397 | + | 150 | 100.0 | 0.11 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (100), E7 (0) | 100.0 |
- 2 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 6092 | chr11 | 114120506 | 114120975 | wgEncodeRegDnaseClusteredV2 | 4 | score:195, srow:235405 |
- 1 features
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 6092 | cellmeth_recounts | PrEC: 0, LNCaP: 1, DU145: 10 | 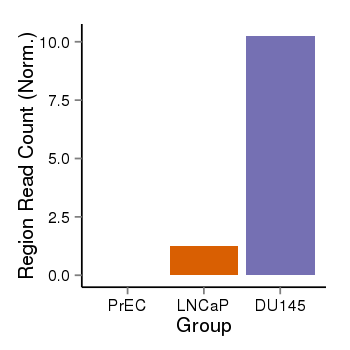 |
| 6092 | cellexp | id=ENSG00000109906, name=ZBTB16, PrEC=0, str=2, LNCaP=1743, str=1050, DU145=10, str=3 | 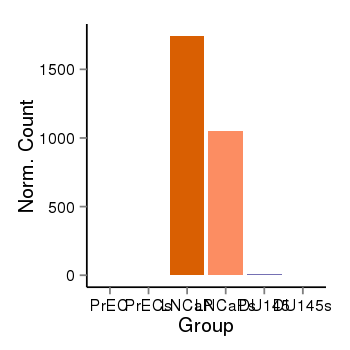 |
| 6092 | cellexp | id=ENSG00000256947, name=RP11-64D24.2, PrEC=0, str=0, LNCaP=1, str=4, DU145=0, str=0 | 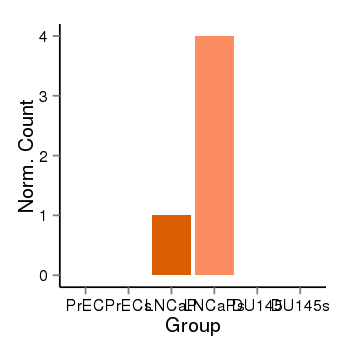 |
| 6092 | tcgaexp | gene=ZBTB16, entrez=7704, pos=chr11:113930431-114121397(+), B=1023, L=1051, M=1266, H=1244 | 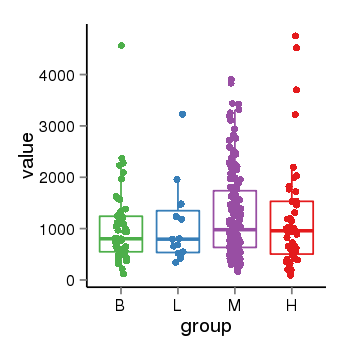 |