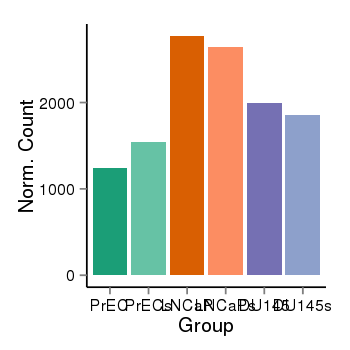DMR 688 chr1:246737151-246737350 (200bp)
| DMR Stats | |
|---|---|
| Position | chr1:246737151-246737350 (View on UCSC) |
| Width | 200bp |
| Genomic Context | intron |
| Nearest Genes | CNST (0bp away) |
| ANODEV p-value | 0.00563836318812 |
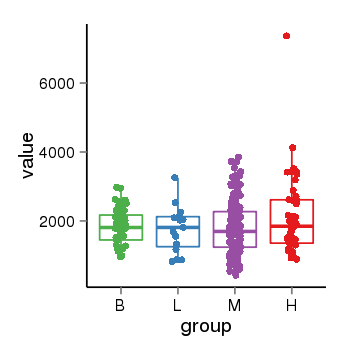
| Frequencies | Benign: 0.0 (0.0%), Low: 0.0 (0.0%), High: 0.0 (0.0 %) |
| Frequent? |
Region Count Boxplot

Region Overview

Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 688 | CNST | 2803 | uc001ibo.4 | chr1 | 246729639 | 246811544 | + | 200 | 100.0 | 0.18 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0), I8 (0), E9 (0) | 0.0 |
| 688 | CNST | 2803 | uc001ibp.3 | chr1 | 246729639 | 246831884 | + | 200 | 100.0 | 0.3 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0), I8 (0), E9 (0), I9 (0), E10 (0), I10 (0), E11 (0) | 0.0 |
- 2 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 688 | chr1 | 246736801 | 246737210 | wgEncodeRegDnaseClusteredV2 | 20 | score:289, srow:119096 |
| 688 | chr1 | 246736913 | 246737282 | wgEncodeRegTfbsClusteredV3 | RUNX3 | score:256, expCount:1, expNums:109, expScores:256, srow:435087 |
- 2 featuress