DMR 690 chr1:246795501-246795650 (150bp)
| DMR Stats | |
|---|---|
| Position | chr1:246795501-246795650 (View on UCSC) |
| Width | 150bp |
| Genomic Context | intron |
| Nearest Genes | CNST (0bp away) |
| ANODEV p-value | 0.0333969371853 |
| Frequencies | Benign: 7.0 (100.0%), Low: 6.0 (100.0%), High: 9.0 (100.0 %) |
| Frequent? |
Region Count Boxplot

Region Overview

Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 690 | CNST | 2803 | uc001ibo.4 | chr1 | 246729639 | 246811544 | + | 150 | 100.0 | 0.19 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (100), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0), I8 (0), E9 (0) | 0.0 |
| 690 | CNST | 2803 | uc001ibp.3 | chr1 | 246729639 | 246831884 | + | 150 | 100.0 | 0.31 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (100), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0), I8 (0), E9 (0), I9 (0), E10 (0), I10 (0), E11 (0) | 0.0 |
- 2 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 690 | chr1 | 246795508 | 246795537 | phastConsElements100way | lod=69 | score:413, srow:860403 |
| 690 | chr1 | 246795550 | 246795785 | phastConsElements100way | lod=838 | score:660, srow:860404 |
- 2 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 690 | cellmeth_recounts | PrEC: 17, LNCaP: 0, DU145: 6 |  |
| 690 | cellmeth_diffreps_PrECvsLNCaP | A=26, B=0, Length=380, Event=Down, log2FC=-Inf, padj=1.35542249872291e-07 |  |
| 690 | cellmeth_diffreps_PrECvsDU145 | A=26, B=5, Length=300, Event=Down, log2FC=-2.38, padj=0.00290451814714991 |  |
| 690 | cellexp | id=ENSG00000162852, name=CNST, PrEC=1247, str=1541, LNCaP=2771, str=2643, DU145=1998, str=1854 | 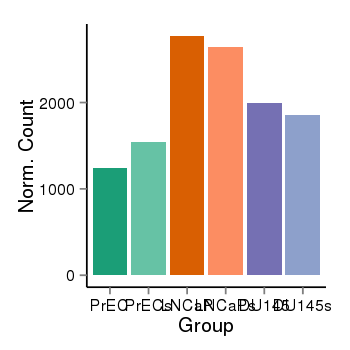 |
| 690 | cellexp | id=ENSG00000228955, name=RP11-452J6.2, PrEC=0, str=0, LNCaP=1, str=9, DU145=3, str=3 |  |
| 690 | tcgameth | site:cg04885562, B=0.94, L=0.92, H=0.92 |  |
| 690 | tcgaexp | gene=CNST, entrez=163882, pos=chr1:246729639-246831884(+), B=1838, L=1764, M=1825, H=2136 | 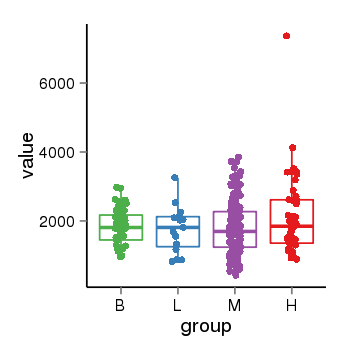 |