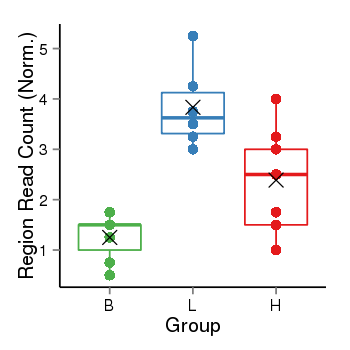
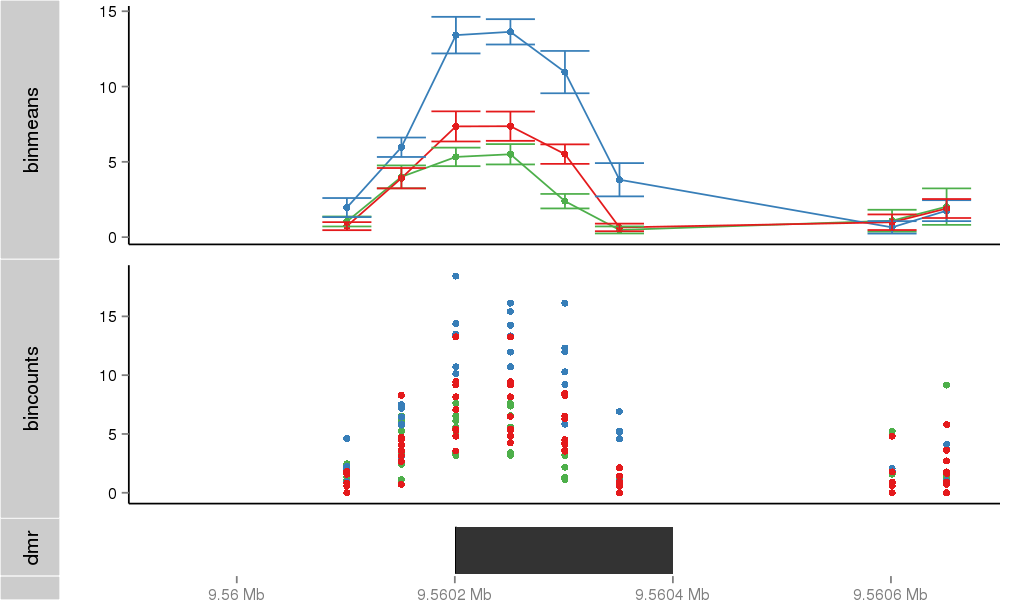
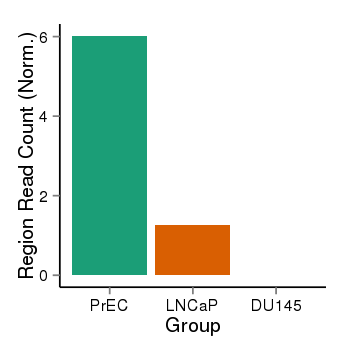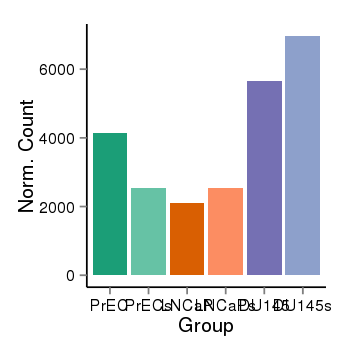| 797 | ITGB1BP1 | 16001 | uc002qzj.3 | chr2 | 9545815 | 9563643 | - | 200 | 100.0 | 0.05 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0) | 0.0 |
| 797 | ITGB1BP1 | 16001 | uc002qzk.3 | chr2 | 9545815 | 9563643 | - | 200 | 100.0 | 0.07 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0) | 0.0 |
| 797 | ITGB1BP1 | 16001 | uc002qzl.3 | chr2 | 9545815 | 9563643 | - | 200 | 100.0 | 0.05 | 1 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0) | 0.0 |
| 797 | ITGB1BP1 | 16001 | uc002qzm.3 | chr2 | 9545815 | 9563643 | - | 200 | 100.0 | 0.04 | 1 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0) | 0.0 |
| 797 | ITGB1BP1 | 16001 | uc002qzn.1 | chr2 | 9546790 | 9563379 | - | 200 | 100.0 | 0.07 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0) | 0.0 |
| 797 | ITGB1BP1 | 16001 | uc010yiy.2 | chr2 | 9545815 | 9563643 | - | 200 | 100.0 | 0.04 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0) | 0.0 |