DMR 897 chr2:39186501-39186700 (200bp)
| DMR Stats | |
|---|---|
| Position | chr2:39186501-39186700 (View on UCSC) |
| Width | 200bp |
| Genomic Context | 3' end |
| Nearest Genes | ARHGEF33, LOC375196 (0bp away) |
| ANODEV p-value | 0.00107099413833 |
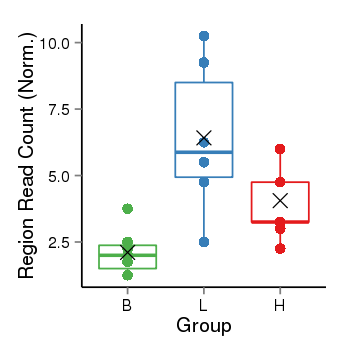
| Frequencies | Benign: 0.0 (0.0%), Low: 5.0 (83.33%), High: 7.0 (77.78 %) |
| Frequent? |
Region Count Boxplot
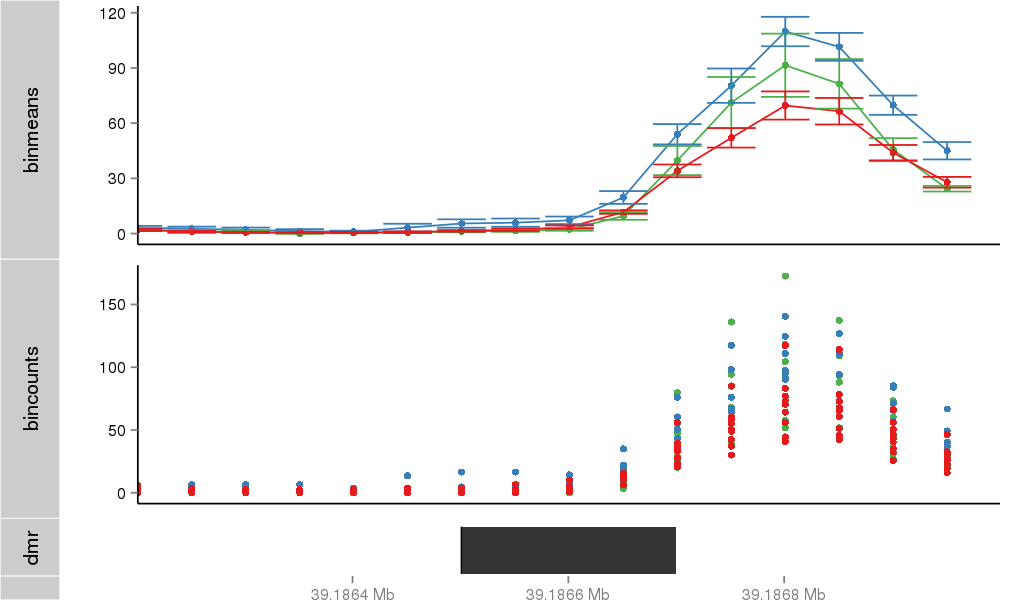
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 897 | ARHGEF33 | 16250 | uc021vgd.1 | chr2 | 39146504 | 39202590 | + | 200 | 100.0 | 0.15 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0), I8 (0), E9 (0), I9 (0), E10 (0), I10 (0), E11 (0), I11 (0), E12 (0), I12 (0), E13 (0), I13 (100), E14 (0), I14 (0), E15 (0), I15 (0), E16 (0) | 0.0 |
| 897 | LOC375196 | 16251 | uc021vge.1 | chr2 | 39186429 | 39187485 | - | 200 | 100.0 | 0.15 | 1 | 0.0 | E1 (100) | 100.0 |
- 2 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 897 | chr2 | 39186566 | 39186755 | wgEncodeRegDnaseClusteredV2 | 5 | score:187, srow:614187 |
| 897 | chr2 | 39184778 | 39186777 | cpgShore | — | values(x.gr)[overs$srow, ]:26789 |
- 2 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 897 | cellmeth_recounts | PrEC: 13, LNCaP: 74, DU145: 13 | 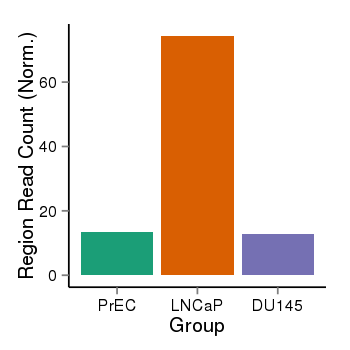 |
| 897 | cellmeth_diffreps_PrECvsLNCaP | A=303, B=2294, Length=1920, Event=Up, log2FC=2.92, padj=0 | 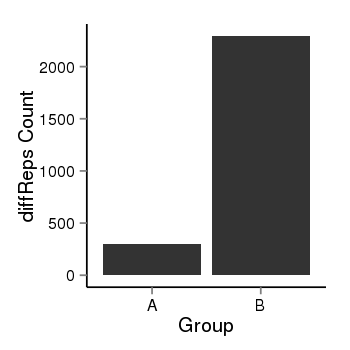 |
| 897 | cellmeth_diffreps_PrECvsDU145 | A=182, B=26, Length=500, Event=Down, log2FC=-2.81, padj=0 | 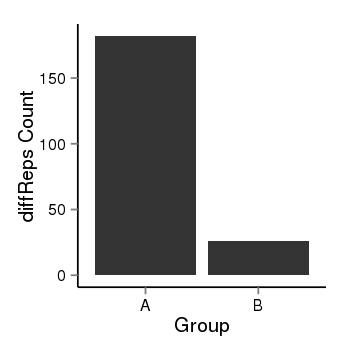 |
| 897 | cellmeth_diffreps_LNCaPvsDU145 | A=2452.39, B=64.54, Length=1940, Event=Down, log2FC=-5.25, padj=0 | 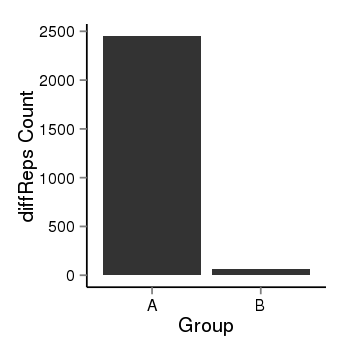 |
| 897 | cellexp | id=ENSG00000214694, name=ARHGEF33, PrEC=3, str=25, LNCaP=2, str=0, DU145=85, str=93 | 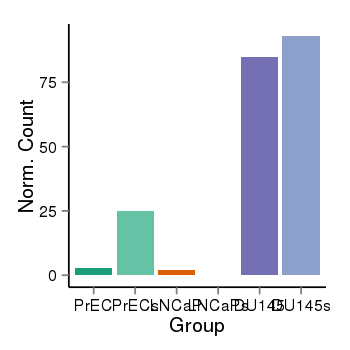 |
| 897 | tcgaexp | gene=ARHGEF33, entrez=100271715, pos=chr2:39146504-39202590(+), B=36, L=24, M=23, H=22 | 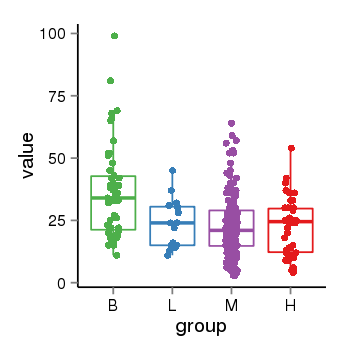 |