DMR 94 chr1:9320051-9320250 (200bp)
| DMR Stats | |
|---|---|
| Position | chr1:9320051-9320250 (View on UCSC) |
| Width | 200bp |
| Genomic Context | intron |
| Nearest Genes | H6PD (0bp away) |
| ANODEV p-value | 0.0112057575451 |
| Frequencies | Benign: 7.0 (100.0%), Low: 6.0 (100.0%), High: 9.0 (100.0 %) |
| Frequent? |
Region Count Boxplot

Region Overview

Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 94 | H6PD | 181 | uc001apt.3 | chr1 | 9294863 | 9331394 | + | 200 | 100.0 | 0.27 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (100), E4 (0), I4 (0), E5 (0) | 0.0 |
- 1 genes
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 94 | chr1 | 9318819 | 9320361 | phastConsElements100way | lod=947 | score:672, srow:33745 |
| 94 | chr1 | 9320203 | 9322202 | cpgShelf | — | values(x.gr)[overs$srow, ]:451 |
- 2 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 94 | cellmeth_recounts | PrEC: 221, LNCaP: 31, DU145: 22 |  |
| 94 | cellmeth_diffreps_PrECvsLNCaP | A=429, B=50, Length=840, Event=Down, log2FC=-3.1, padj=0 |  |
| 94 | cellmeth_diffreps_PrECvsDU145 | A=429, B=30, Length=840, Event=Down, log2FC=-3.84, padj=0 |  |
| 94 | cellexp | id=ENSG00000049239, name=H6PD, PrEC=3604, str=4527, LNCaP=5612, str=4969, DU145=3444, str=3028 |  |
| 94 | tcgaexp | gene=H6PD, entrez=9563, pos=chr1:9294863-9331394(+), B=7314, L=6689, M=6614, H=5703 |  |
| 94 | tcgaexp | gene=SRSF10, entrez=10772, pos=chr1:36273-24306953(-), B=3487, L=3388, M=3432, H=4019 | 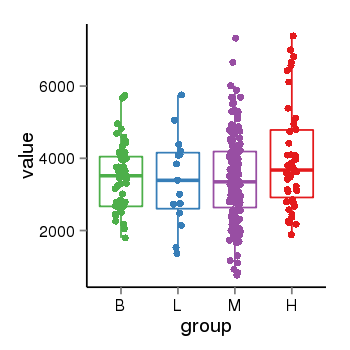 |
| 94 | tcgaexp | gene=LOC100133331, entrez=100133331, pos=chr1:322037-180753553(-), B=268, L=328, M=271, H=305 | 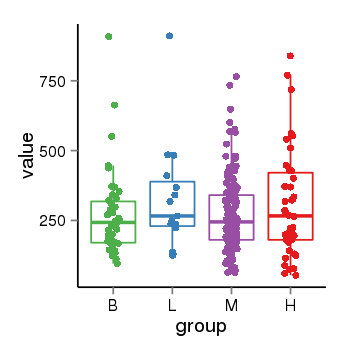 |