DMR 1293 chr2:176965201-176965350 (150bp)
| DMR Stats | |
|---|---|
| Position | chr2:176965201-176965350 (View on UCSC) |
| Width | 150bp |
| Genomic Context | 3' end |
| Nearest Genes | HOXD12 (0bp away) |
| ANODEV p-value | 0.0135954910277 |
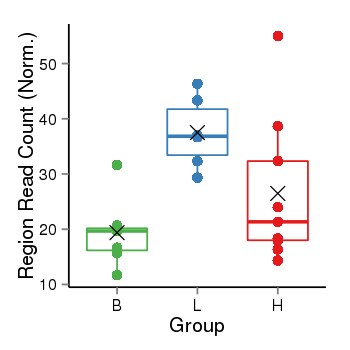
| Frequencies | Benign: 7.0 (100.0%), Low: 6.0 (100.0%), High: 9.0 (100.0 %) |
| Frequent? |
Region Count Boxplot
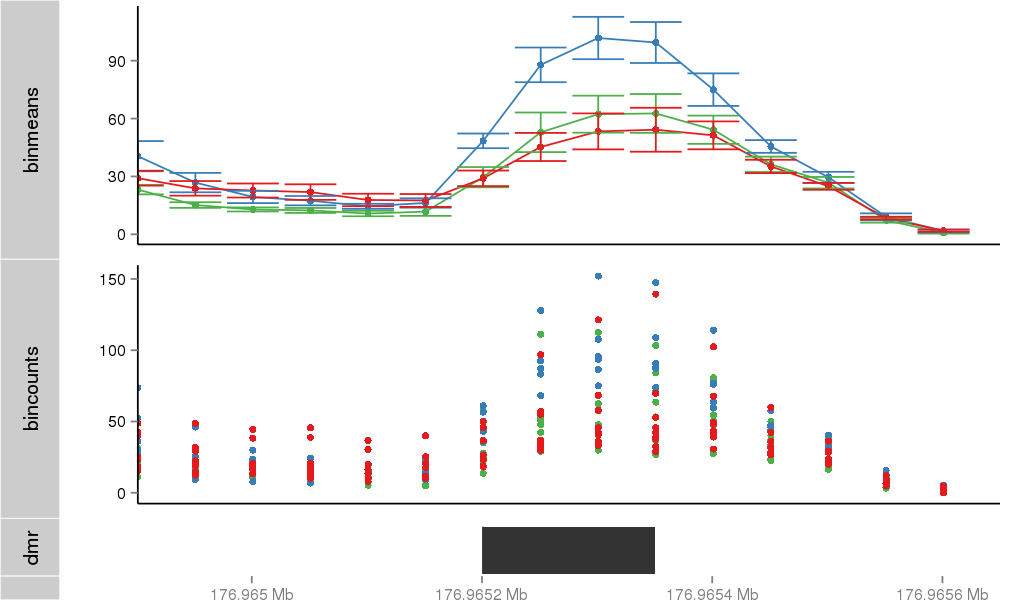
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1293 | HOXD12 | 17298 | uc010zev.1 | chr2 | 176964530 | 176965488 | + | 150 | 100.0 | 0.07 | 0 | 0.0 | E1 (0), I1 (32.67), E2 (67.33) | 100.0 |
| 1293 | HOXD12 | 17298 | uc021vsp.1 | chr2 | 176964530 | 176965488 | + | 150 | 100.0 | 0.11 | 0 | 0.0 | E1 (100) | 100.0 |
- 2 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 1293 | chr2 | 176965321 | 176965340 | phastConsElements100way | lod=248 | score:539, srow:4969120 |
| 1293 | chr2 | 176965343 | 176965361 | phastConsElements100way | lod=223 | score:529, srow:4969121 |
| 1293 | chr2 | 176964063 | 176965509 | cpgIsland | CpG: 118 | length:1447, cpgNum:118, gcNum:909, perCpg:16.3, perGc:62.8, obsExp:0.83, srow:15754 |
- Previous
- Page 2 of 2
- 3 of 28 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 1293 | cellmeth_recounts | PrEC: 237, LNCaP: 186, DU145: 8 | 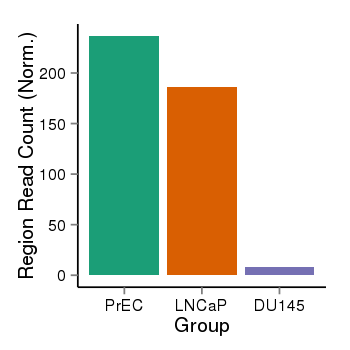 |
| 1293 | cellmeth_diffreps_PrECvsLNCaP | A=1457, B=739, Length=1680, Event=Down, log2FC=-0.98, padj=0 | 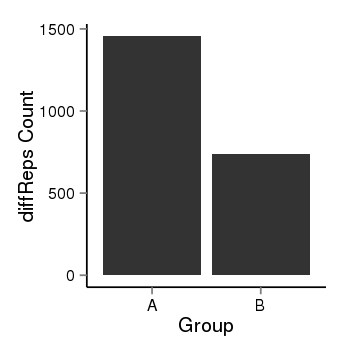 |
| 1293 | cellmeth_diffreps_PrECvsDU145 | A=1460, B=53, Length=1800, Event=Down, log2FC=-4.78, padj=0 | 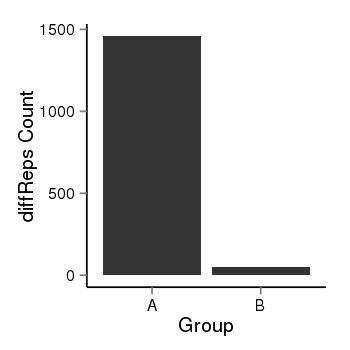 |
| 1293 | cellmeth_diffreps_LNCaPvsDU145 | A=799.65, B=49.58, Length=1780, Event=Down, log2FC=-4.01, padj=0 | 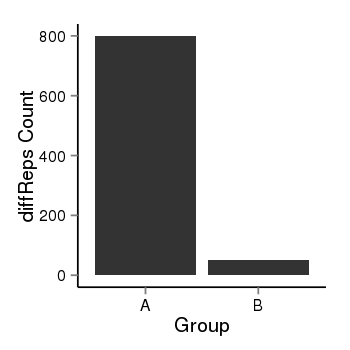 |
| 1293 | cellexp | id=ENSG00000170178, name=HOXD12, PrEC=3, str=4, LNCaP=0, str=0, DU145=0, str=0 | 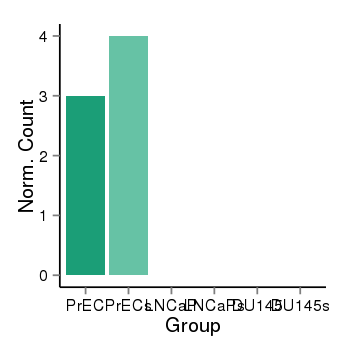 |
| 1293 | tcgaexp | gene=HOXD12, entrez=3238, pos=chr2:176964530-176965488(+), B=12, L=2, M=4, H=3 | 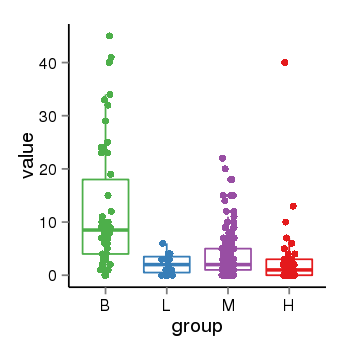 |