DMR 21046 chr16:3433251-3433400 (150bp)
| DMR Stats | |
|---|---|
| Position | chr16:3433251-3433400 (View on UCSC) |
| Width | 150bp |
| Genomic Context | 3' end |
| Nearest Genes | ZSCAN32 (0bp away) |
| ANODEV p-value | 0.0140969238346 |
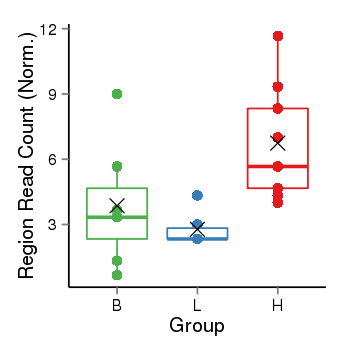
| Frequencies | Benign: 5.0 (71.43%), Low: 1.0 (16.67%), High: 9.0 (100.0 %) |
| Frequent? |
Region Count Boxplot
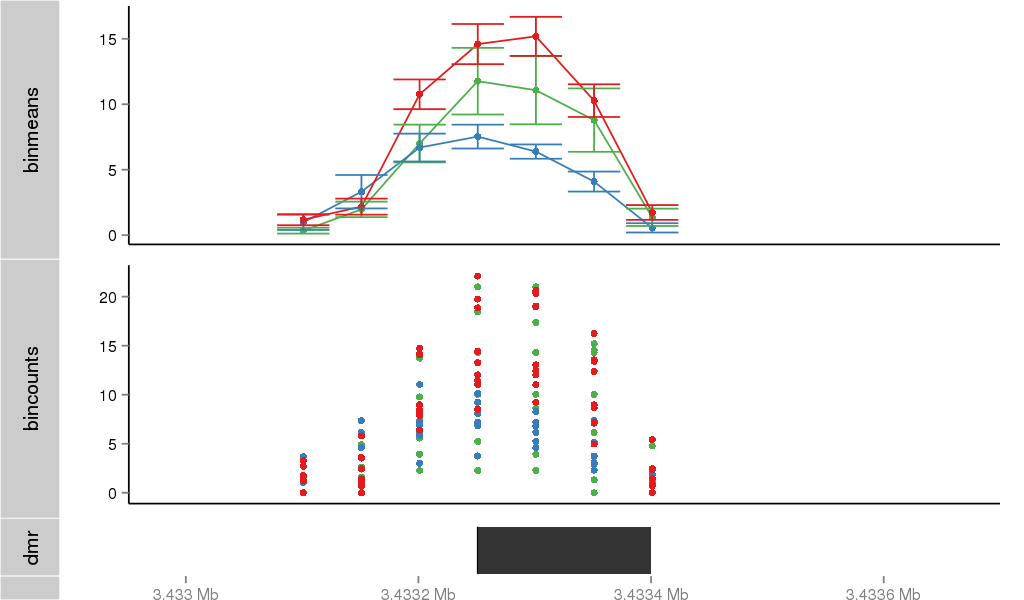
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 21046 | ZSCAN32 | 10873 | uc002cux.4 | chr16 | 3432085 | 3451013 | - | 150 | 100.0 | 1.02 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (100) | 0.0 |
| 21046 | ZSCAN32 | 10873 | uc002cuy.4 | chr16 | 3432085 | 3451013 | - | 150 | 100.0 | 1.02 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (100) | 0.0 |
| 21046 | ZSCAN32 | 10873 | uc002cuz.3 | chr16 | 3432085 | 3451025 | - | 150 | 100.0 | 0.02 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (100) | 0.0 |
| 21046 | ZSCAN32 | 10873 | uc010uwx.2 | chr16 | 3432085 | 3451013 | - | 150 | 100.0 | 0.04 | 0 | 0.0 | E1 (0), I1 (0), E2 (0), I2 (0), E3 (100) | 0.0 |
- 4 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 21046 | chr16 | 3433378 | 3433379 | phastConsElements100way | lod=12 | score:240, srow:3234369 |
- Previous
- Page 2 of 2
- 1 of 26 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 21046 | cellmeth_recounts | PrEC: 5, LNCaP: 0, DU145: 0 | 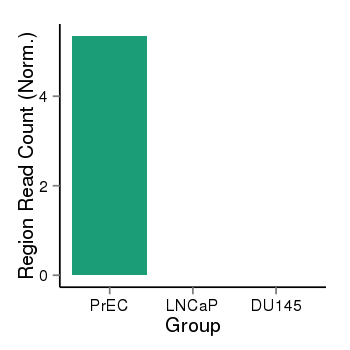 |
| 21046 | cellexp | id=ENSG00000140987, name=ZSCAN32, PrEC=856, str=912, LNCaP=1050, str=1163, DU145=796, str=819 | 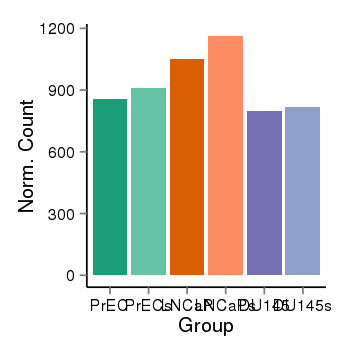 |
| 21046 | cellexp | id=ENSG00000262621, name=NAA60, PrEC=5, str=2, LNCaP=0, str=1, DU145=0, str=0 | 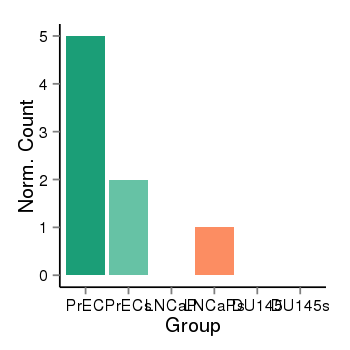 |
| 21046 | tcgaexp | gene=ZSCAN32, entrez=54925, pos=chr16:3432085-3451025(-), B=799, L=729, M=781, H=854 | 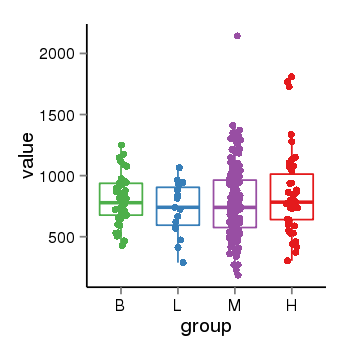 |