DMR 33558 chr16:8806501-8807000 (500bp)
| DMR Stats | |
|---|---|
| Position | chr16:8806501-8807000 (View on UCSC) |
| Width | 500bp |
| Genomic Context | promoter |
| Nearest Genes | ABAT (0bp away) |
| ANODEV p-value | 2.10773639929e-07 |
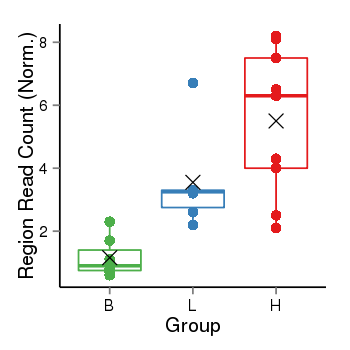
| Frequencies | Benign: 0.0 (0.0%), Low: 4.0 (66.67%), High: 7.0 (77.78 %) |
| Frequent? |
Region Count Boxplot
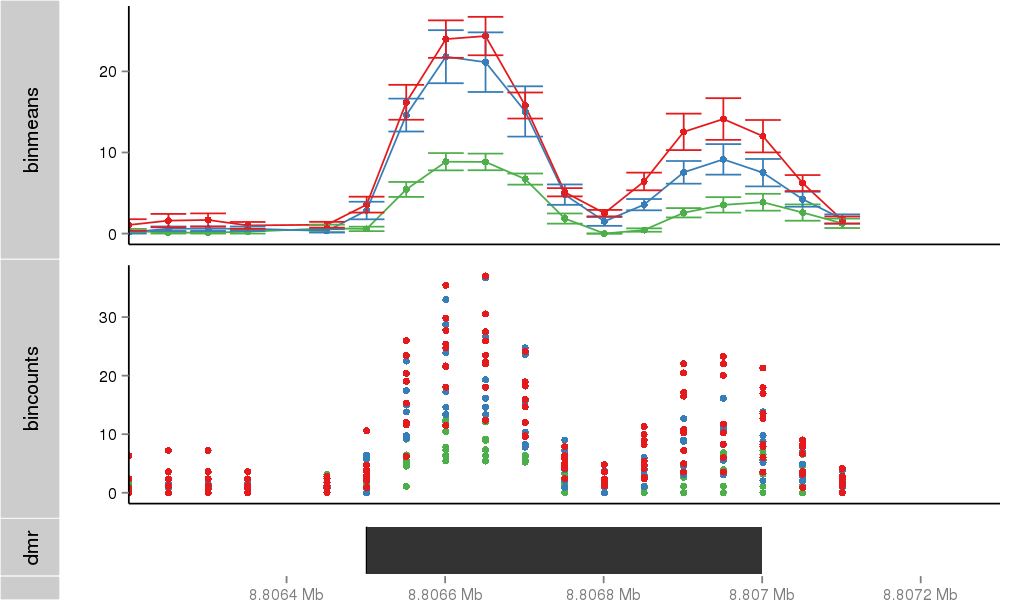
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 33558 | ABAT | 10923 | uc002czc.4 | chr16 | 8768444 | 8878432 | + | 500 | 100.0 | 2.55 | 0 | 0.0 | E1 (0), I1 (100), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0), I8 (0), E9 (0), I9 (0), E10 (0), I10 (0), E11 (0), I11 (0), E12 (0), I12 (0), E13 (0), I13 (0), E14 (0), I14 (0), E15 (0), I15 (0), E16 (0) | 0.0 |
| 33558 | ABAT | 10923 | uc002czd.4 | chr16 | 8806826 | 8878432 | + | 175 | 35.0 | 0.36 | 0 | 100.0 | E1 (35), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0), I8 (0), E9 (0), I9 (0), E10 (0), I10 (0), E11 (0), I11 (0), E12 (0), I12 (0), E13 (0), I13 (0), E14 (0), I14 (0), E15 (0), I15 (0), E16 (0) | 0.0 |
| 33558 | ABAT | 10923 | uc010buh.3 | chr16 | 8806826 | 8878432 | + | 175 | 35.0 | 0.6 | 0 | 100.0 | E1 (35), I1 (0), E2 (0), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0), I6 (0), E7 (0), I7 (0), E8 (0), I8 (0), E9 (0), I9 (0), E10 (0), I10 (0), E11 (0), I11 (0), E12 (0), I12 (0), E13 (0), I13 (0), E14 (0), I14 (0), E15 (0) | 0.0 |
- 3 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 33558 | chr16 | 8806572 | 8806582 | phastConsElements100way | lod=40 | score:359, srow:3253837 |
| 33558 | chr16 | 8806596 | 8806652 | phastConsElements100way | lod=312 | score:562, srow:3253838 |
| 33558 | chr16 | 8806656 | 8806673 | phastConsElements100way | lod=101 | score:450, srow:3253839 |
| 33558 | chr16 | 8806676 | 8806689 | phastConsElements100way | lod=56 | score:392, srow:3253840 |
| 33558 | chr16 | 8806693 | 8806704 | phastConsElements100way | lod=61 | score:400, srow:3253841 |
| 33558 | chr16 | 8806711 | 8806714 | phastConsElements100way | lod=24 | score:308, srow:3253842 |
| 33558 | chr16 | 8806719 | 8806725 | phastConsElements100way | lod=21 | score:295, srow:3253843 |
| 33558 | chr16 | 8806733 | 8806735 | phastConsElements100way | lod=13 | score:247, srow:3253844 |
| 33558 | chr16 | 8806747 | 8806752 | phastConsElements100way | lod=14 | score:255, srow:3253845 |
| 33558 | chr16 | 8806757 | 8806779 | phastConsElements100way | lod=105 | score:454, srow:3253846 |
- Previous
- Page 2 of 2
- 10 of 35 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 33558 | cellmeth_recounts | PrEC: 12, LNCaP: 0, DU145: 5 | 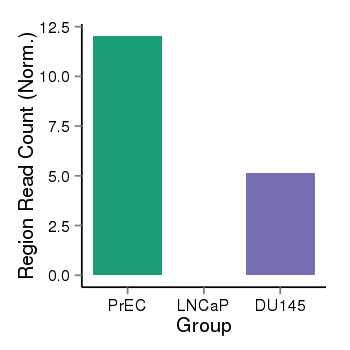 |
| 33558 | cellmeth_diffreps_PrECvsLNCaP | A=12, B=0, Length=300, Event=Down, log2FC=-Inf, padj=0.00157949948552458 | 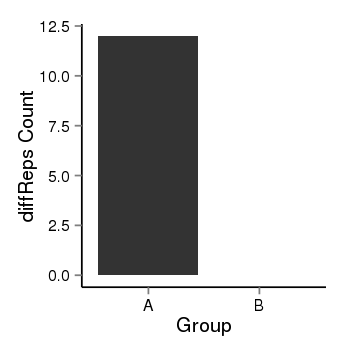 |
| 33558 | cellexp | id=ENSG00000183044, name=ABAT, PrEC=1039, str=1896, LNCaP=2957, str=4501, DU145=276, str=56 | 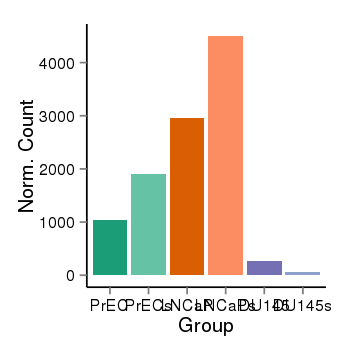 |
| 33558 | tcgameth | site:cg00220102, B=0.51, L=0.74, H=0.73 | 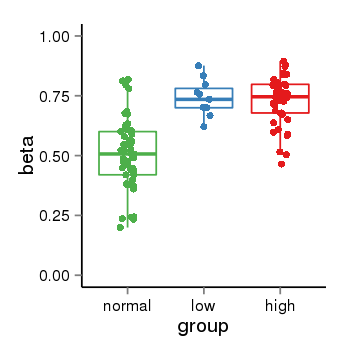 |
| 33558 | tcgameth | site:cg00231389, B=0.32, L=0.56, H=0.56 | 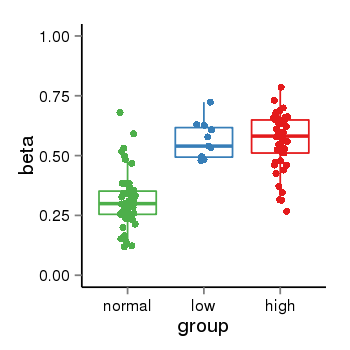 |
| 33558 | tcgameth | site:cg01881182, B=0.63, L=0.81, H=0.8 | 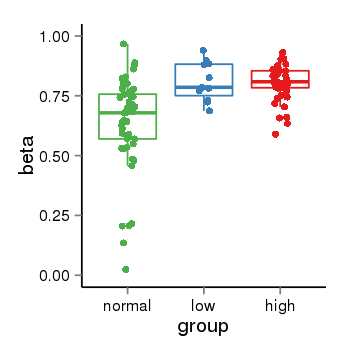 |
| 33558 | tcgameth | site:cg08241330, B=0.57, L=0.73, H=0.73 | 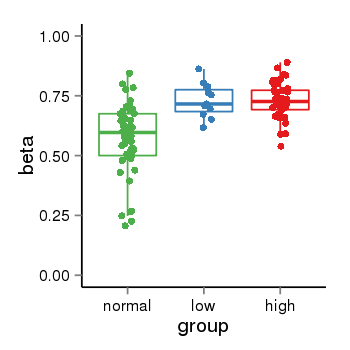 |
| 33558 | tcgameth | site:cg08834902, B=0.56, L=0.72, H=0.72 | 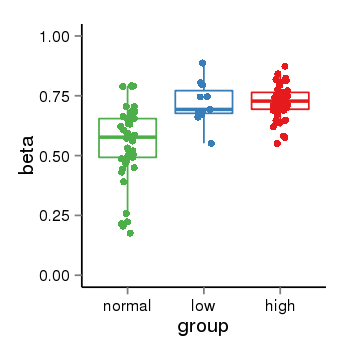 |
| 33558 | tcgameth | site:cg16586594, B=0.51, L=0.72, H=0.72 | 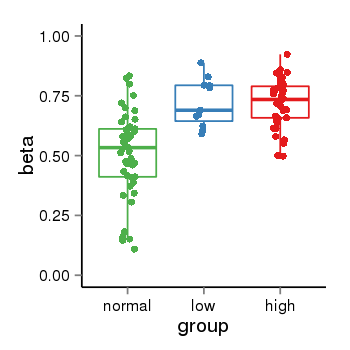 |
| 33558 | tcgameth | site:cg26778345, B=0.35, L=0.65, H=0.62 | 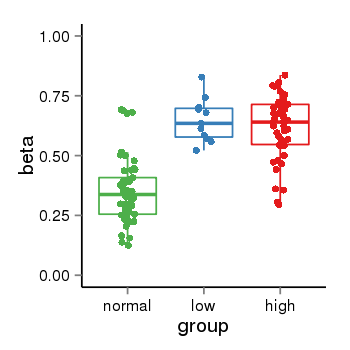 |
| 33558 | tcgaexp | gene=ABAT, entrez=18, pos=chr16:8768444-8878432(+), B=5178, L=7494, M=7026, H=5106 | 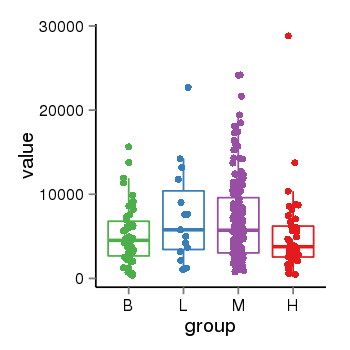 |