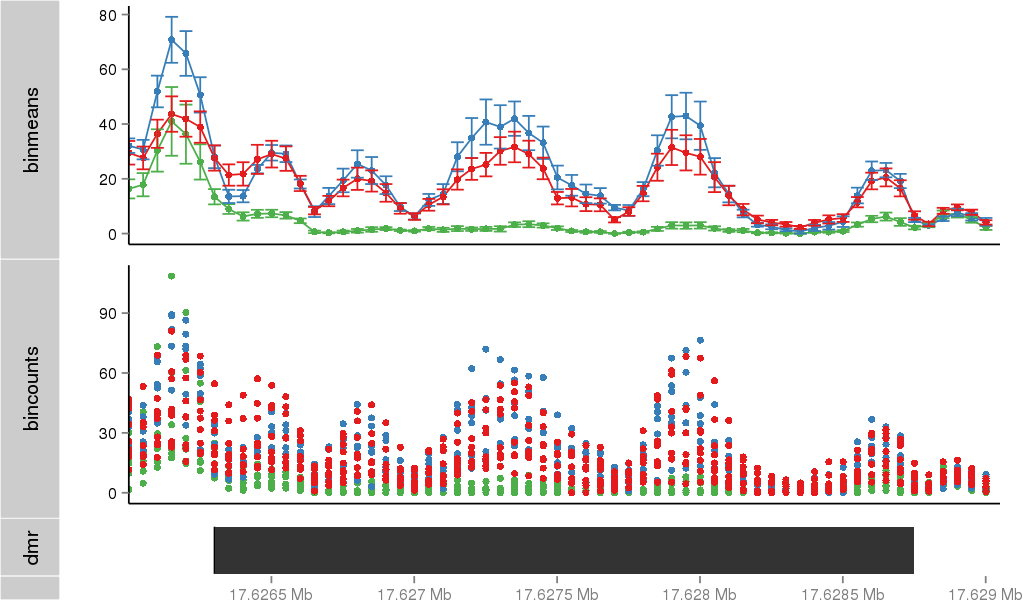
DMR 34060 chr17:17626301-17628750 (2450bp)
| DMR Stats | |
|---|---|
| Position | chr17:17626301-17628750 (View on UCSC) |
| Width | 2450bp |
| Genomic Context | exon |
| Nearest Genes | RAI1 (0bp away) |
| ANODEV p-value | 7.14529637478e-09 |
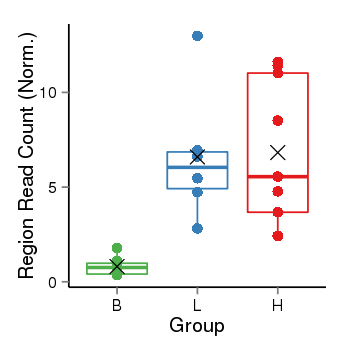
| Frequencies | Benign: 0.0 (0.0%), Low: 5.0 (83.33%), High: 7.0 (77.78 %) |
| Frequent? |
Region Count Boxplot
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 34060 | RAI1 | 12265 | uc002grm.3 | chr17 | 17584787 | 17714765 | + | 2450 | 100.0 | 0.12 | 0 | 0.0 | E1 (0), I1 (42.49), E2 (5.39), I2 (52.16), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0) | 0.0 |
- 1 genes
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 34060 | chr17 | 17628416 | 17628920 | wgEncodeRegTfbsClusteredV3 | POLR2A | score:501, expCount:4, expNums:206,461,632,633, expScores:148,225,501,255, srow:1645159 |
| 34060 | chr17 | 17628507 | 17628810 | wgEncodeRegTfbsClusteredV3 | FOXP2 | score:173, expCount:1, expNums:251, expScores:173, srow:1645160 |
| 34060 | chr17 | 17628568 | 17628757 | wgEncodeRegTfbsClusteredV3 | MYC | score:108, expCount:1, expNums:617, expScores:108, srow:1645161 |
| 34060 | chr17 | 17626373 | 17626385 | tfbsConsSites | V$CHOP_01 | score:833, zScore:1.78, srow:2106579 |
| 34060 | chr17 | 17626430 | 17626440 | tfbsConsSites | V$FOXO4_01 | score:909, zScore:1.68, srow:2106580 |
| 34060 | chr17 | 17626525 | 17626536 | tfbsConsSites | V$EGR1_01 | score:955, zScore:3.74, srow:2106581 |
| 34060 | chr17 | 17626525 | 17626536 | tfbsConsSites | V$EGR2_01 | score:965, zScore:3.84, srow:2106582 |
| 34060 | chr17 | 17626525 | 17626536 | tfbsConsSites | V$EGR3_01 | score:974, zScore:3.93, srow:2106583 |
| 34060 | chr17 | 17626525 | 17626536 | tfbsConsSites | V$NGFIC_01 | score:925, zScore:3.24, srow:2106584 |
| 34060 | chr17 | 17626559 | 17626574 | tfbsConsSites | V$FREAC2_01 | score:882, zScore:2.95, srow:2106585 |
| 34060 | chr17 | 17626561 | 17626572 | tfbsConsSites | V$CDP_01 | score:811, zScore:2.36, srow:2106586 |
| 34060 | chr17 | 17626562 | 17626573 | tfbsConsSites | V$HFH1_01 | score:901, zScore:2.36, srow:2106587 |
| 34060 | chr17 | 17626563 | 17626572 | tfbsConsSites | V$FOXO1_01 | score:952, zScore:1.72, srow:2106588 |
| 34060 | chr17 | 17626564 | 17626574 | tfbsConsSites | V$FOXO4_01 | score:934, zScore:2.09, srow:2106589 |
| 34060 | chr17 | 17626565 | 17626574 | tfbsConsSites | V$SOX5_01 | score:957, zScore:2.24, srow:2106590 |
| 34060 | chr17 | 17626675 | 17626698 | tfbsConsSites | V$BRACH_01 | score:705, zScore:2.21, srow:2106591 |
| 34060 | chr17 | 17626746 | 17626757 | tfbsConsSites | V$PAX4_03 | score:906, zScore:1.67, srow:2106592 |
| 34060 | chr17 | 17626827 | 17626838 | tfbsConsSites | V$EGR2_01 | score:848, zScore:2.01, srow:2106593 |
| 34060 | chr17 | 17626827 | 17626838 | tfbsConsSites | V$EGR3_01 | score:840, zScore:1.97, srow:2106594 |
| 34060 | chr17 | 17626949 | 17626961 | tfbsConsSites | V$PAX3_01 | score:798, zScore:1.71, srow:2106595 |
| 34060 | chr17 | 17627002 | 17627011 | tfbsConsSites | V$SP1_01 | score:929, zScore:1.75, srow:2106596 |
| 34060 | chr17 | 17627005 | 17627017 | tfbsConsSites | V$MAZR_01 | score:871, zScore:1.89, srow:2106597 |
| 34060 | chr17 | 17627023 | 17627041 | tfbsConsSites | V$PAX2_01 | score:775, zScore:1.8, srow:2106598 |
| 34060 | chr17 | 17627036 | 17627051 | tfbsConsSites | V$AHRARNT_01 | score:837, zScore:1.82, srow:2106599 |
| 34060 | chr17 | 17627057 | 17627086 | tfbsConsSites | V$HOX13_01 | score:738, zScore:2.27, srow:2106600 |
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 34060 | cellmeth_recounts | PrEC: 80, LNCaP: 4849, DU145: 61 | 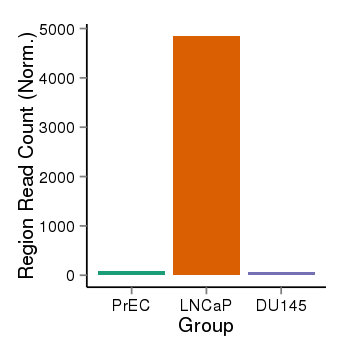 |
| 34060 | cellmeth_diffreps_PrECvsLNCaP | A=583, B=5143, Length=3860, Event=Up, log2FC=3.14, padj=0 | 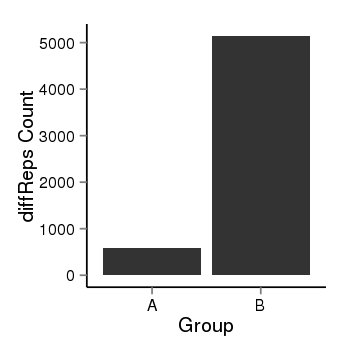 |
| 34060 | cellmeth_diffreps_PrECvsDU145 | A=550, B=23, Length=1340, Event=Down, log2FC=-4.58, padj=0 | 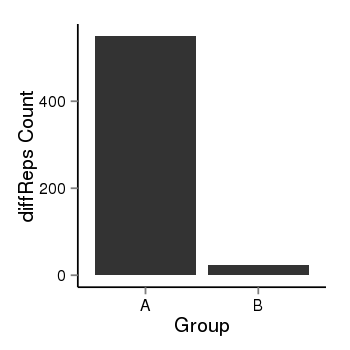 |
| 34060 | cellmeth_diffreps_LNCaPvsDU145 | A=5498.1, B=59.87, Length=3880, Event=Down, log2FC=-6.52, padj=0 | 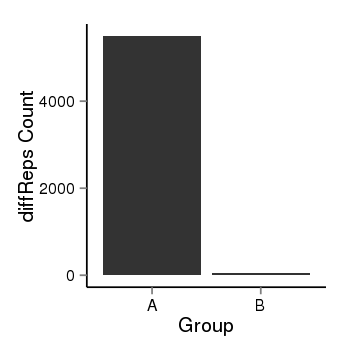 |
| 34060 | cellexp | id=ENSG00000108557, name=RAI1, PrEC=8222, str=4496, LNCaP=5636, str=3754, DU145=1644, str=1097 | 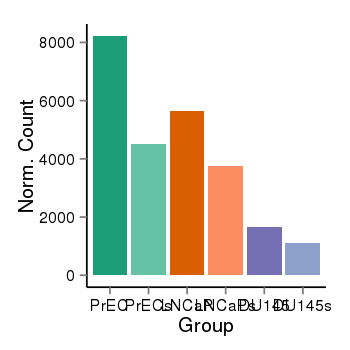 |
| 34060 | tcgameth | site:cg00073010, B=0.15, L=0.59, H=0.54 | 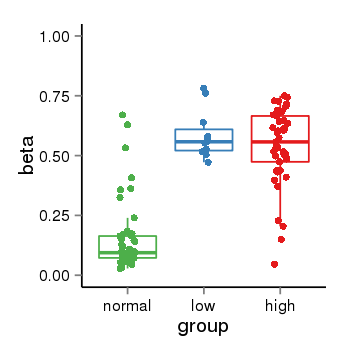 |
| 34060 | tcgameth | site:cg13000649, B=0.12, L=0.54, H=0.48 | 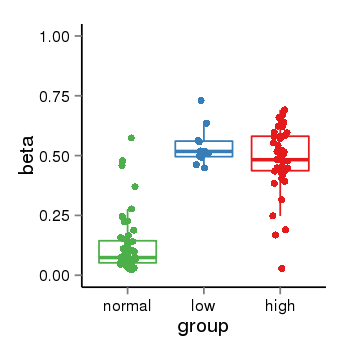 |
| 34060 | tcgameth | site:cg19447962, B=0.35, L=0.78, H=0.71 | 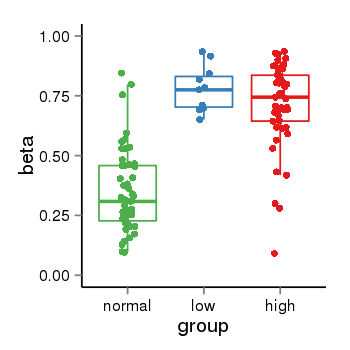 |
| 34060 | tcgaexp | gene=CRHR1, entrez=1394, pos=chr17:954315-43913194(+), B=36, L=21, M=25, H=18 | 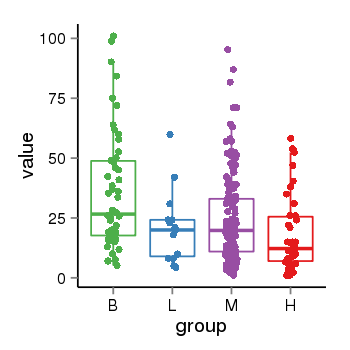 |
| 34060 | tcgaexp | gene=MAPT, entrez=4137, pos=chr17:762281-44105699(+), B=1933, L=2382, M=2487, H=2583 | 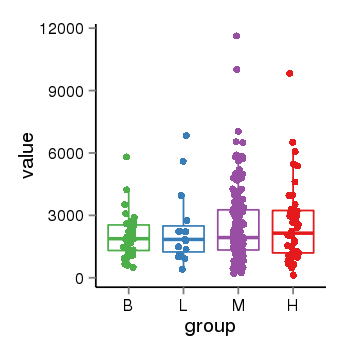 |
| 34060 | tcgaexp | gene=NSF, entrez=4905, pos=chr17:266212-44834828(+), B=3355, L=5678, M=5410, H=5700 | 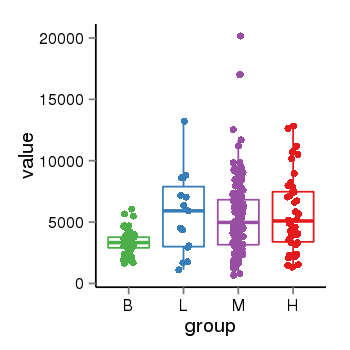 |
| 34060 | tcgaexp | gene=PLEKHM1, entrez=9842, pos=chr17:128328-43568146(-), B=1993, L=1631, M=1589, H=1418 | 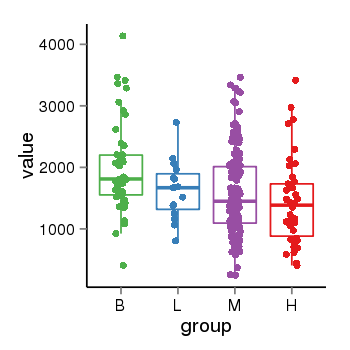 |
| 34060 | tcgaexp | gene=LRRC37A, entrez=9884, pos=chr17:415627-44415160(+), B=924, L=896, M=971, H=940 | 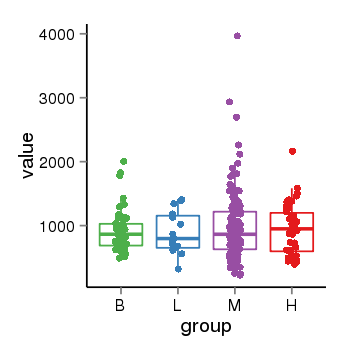 |
| 34060 | tcgaexp | gene=RAI1, entrez=10743, pos=chr17:17584787-17714765(+), B=3085, L=2862, M=2789, H=2953 | 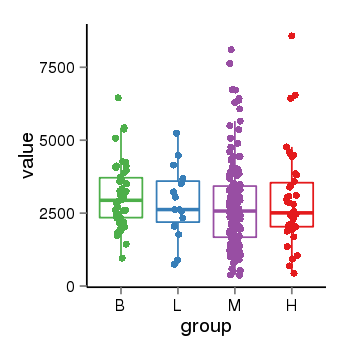 |
| 34060 | tcgaexp | gene=ARL17A, entrez=51326, pos=chr17:195721-44657088(-), B=200, L=195, M=210, H=230 | 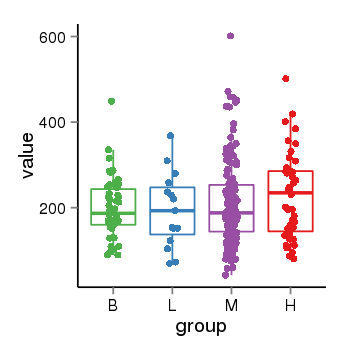 |
| 34060 | tcgaexp | gene=LRRC37A4P, entrez=55073, pos=chr17:197706-43597889(-), B=1015, L=1512, M=1396, H=1450 | 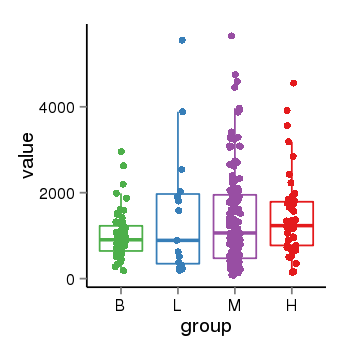 |
| 34060 | tcgaexp | gene=CRHR1-IT1, entrez=147081, pos=chr17:1143726-43723595(+), B=197, L=211, M=215, H=223 | 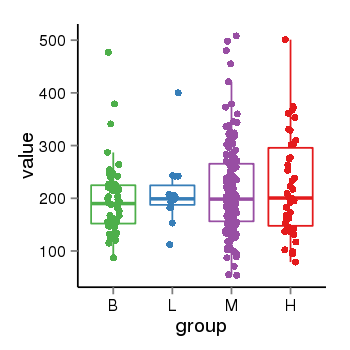 |
| 34060 | tcgaexp | gene=SPPL2C, entrez=162540, pos=chr17:943102-43924438(+), B=0, L=0, M=0, H=0 | 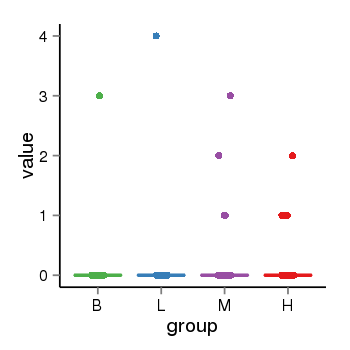 |
| 34060 | tcgaexp | gene=ARHGAP27, entrez=201176, pos=chr17:86404-43511112(-), B=1570, L=1264, M=1273, H=1486 | 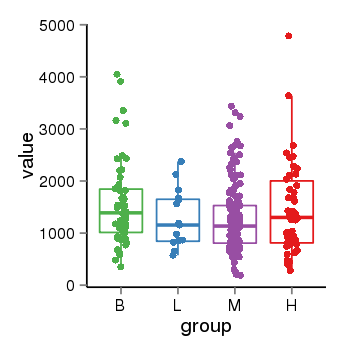 |
| 34060 | tcgaexp | gene=STH, entrez=246744, pos=chr17:790689-44077060(+), B=0, L=0, M=0, H=0 | 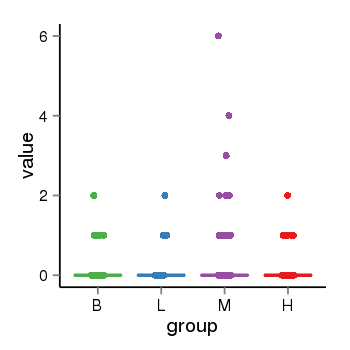 |
| 34060 | tcgaexp | gene=KANSL1, entrez=284058, pos=chr17:563498-44302740(-), B=3141, L=2594, M=2797, H=3017 | 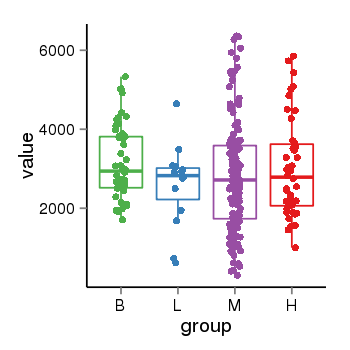 |
| 34060 | tcgaexp | gene=MGC57346, entrez=401884, pos=chr17:1151952-43715329(+), B=601, L=581, M=566, H=598 | 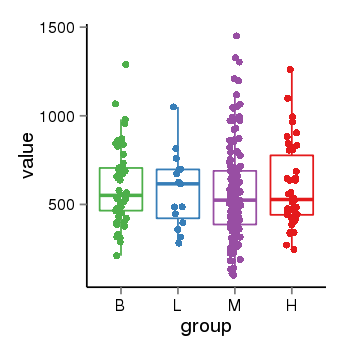 |
| 34060 | tcgaexp | gene=LRRC37A2, entrez=474170, pos=chr17:197706-44633014(+), B=351, L=310, M=361, H=377 | 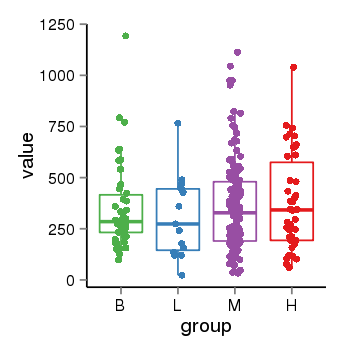 |
| 34060 | tcgaexp | gene=LOC644172, entrez=644172, pos=chr17:1188439-43679748(-), B=84, L=82, M=93, H=90 | 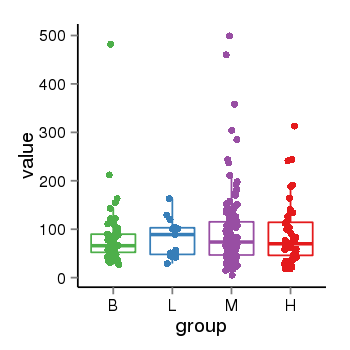 |
| 34060 | tcgaexp | gene=MAPT-AS1, entrez=100128977, pos=chr17:894694-43972879(-), B=1, L=2, M=4, H=2 | 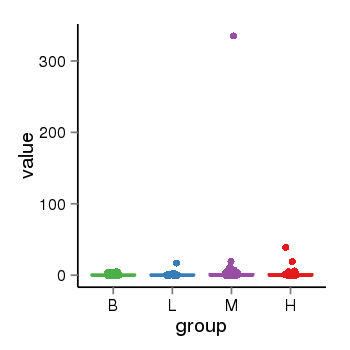 |
| 34060 | tcgaexp | gene=MAPT-IT1, entrez=100130148, pos=chr17:891434-43976164(+), B=5, L=3, M=3, H=4 | 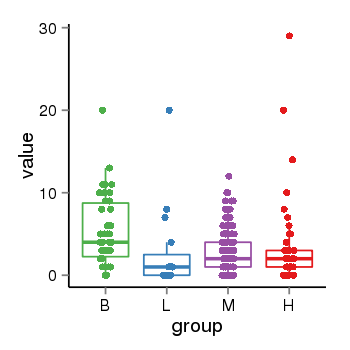 |