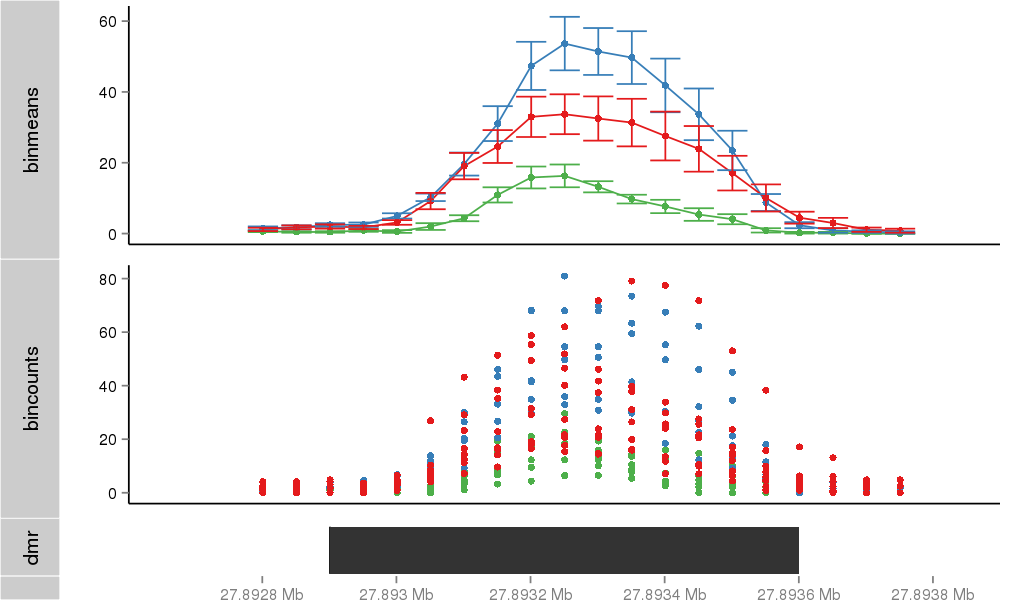
DMR 34110 chr17:27892901-27893600 (700bp)
| DMR Stats | |
|---|---|
| Position | chr17:27892901-27893600 (View on UCSC) |
| Width | 700bp |
| Genomic Context | promoter |
| Nearest Genes | ABHD15 (0bp away) |
| ANODEV p-value | 0.000242374832034 |
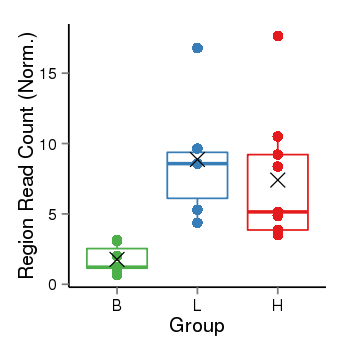
| Frequencies | Benign: 2.0 (28.57%), Low: 6.0 (100.0%), High: 9.0 (100.0 %) |
| Frequent? |
Region Count Boxplot
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 34110 | ABHD15 | 12501 | uc002hed.2 | chr17 | 27887689 | 27894042 | - | 700 | 100.0 | 0.86 | 0 | 8.29 | E1 (71), I1 (29), E2 (0) | 0.0 |
- 1 genes
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 34110 | chr17 | 27893425 | 27894334 | wgEncodeRegTfbsClusteredV3 | E2F1 | score:497, expCount:3, expNums:384,390,572, expScores:346,164,330, srow:1662318 |
| 34110 | chr17 | 27893463 | 27893992 | wgEncodeRegTfbsClusteredV3 | TFAP2A | score:285, expCount:1, expNums:369, expScores:285, srow:1662319 |
| 34110 | chr17 | 27893480 | 27894264 | wgEncodeRegTfbsClusteredV3 | TFAP2C | score:452, expCount:1, expNums:370, expScores:155, srow:1662320 |
| 34110 | chr17 | 27893599 | 27894094 | wgEncodeRegTfbsClusteredV3 | CTCF | score:311, expCount:11, expNums:595,597,604,605,606,607,628,630,631,651,655, expScores:168,120,311,117,148,175,278,148,197,206,111, srow:1662321 |
| 34110 | chr17 | 27893154 | 27893163 | tfbsConsSites | V$P53_02 | score:957, zScore:1.98, srow:2121634 |
| 34110 | chr17 | 27893230 | 27893244 | tfbsConsSites | V$TAXCREB_02 | score:765, zScore:2.03, srow:2121635 |
| 34110 | chr17 | 27893282 | 27893296 | tfbsConsSites | V$E2F_01 | score:816, zScore:2.85, srow:2121636 |
| 34110 | chr17 | 27893298 | 27893309 | tfbsConsSites | V$AP2_Q6 | score:908, zScore:2.17, srow:2121637 |
| 34110 | chr17 | 27893311 | 27893318 | tfbsConsSites | V$STAT3_02 | score:985, zScore:1.64, srow:2121638 |
| 34110 | chr17 | 27893383 | 27893403 | tfbsConsSites | V$PAX4_01 | score:820, zScore:2.06, srow:2121639 |
| 34110 | chr17 | 27893387 | 27893406 | tfbsConsSites | V$YY1_02 | score:775, zScore:1.88, srow:2121640 |
| 34110 | chr17 | 27893393 | 27893413 | tfbsConsSites | V$NRSF_01 | score:729, zScore:2.03, srow:2121641 |
| 34110 | chr17 | 27893406 | 27893425 | tfbsConsSites | V$YY1_02 | score:762, zScore:1.66, srow:2121642 |
| 34110 | chr17 | 27893493 | 27893502 | tfbsConsSites | V$NFKAPPAB_01 | score:902, zScore:1.72, srow:2121643 |
| 34110 | chr17 | 27893557 | 27893576 | tfbsConsSites | V$PPARA_01 | score:757, zScore:2.09, srow:2121644 |
| 34110 | chr17 | 27893109 | 27893109 | cosmic | COSM352646 | srow:489240 |
| 34110 | chr17 | 27893424 | 27893424 | cosmic | COSM560453 | srow:489241 |
| 34110 | chr17 | 27893470 | 27893470 | cosmic | COSM312549 | srow:489242 |
| 34110 | chr17 | 27893483 | 27893483 | cosmic | COSM472506 | srow:489243 |
| 34110 | chr17 | 27893552 | 27893554 | cosmic | COSM1190910 | srow:489244 |
| 34110 | chr17 | 27893568 | 27893568 | cosmic | COSM1679678 | srow:489245 |
| 34110 | chr17 | 27893098 | 27893111 | phastConsElements100way | lod=127 | score:473, srow:3631853 |
| 34110 | chr17 | 27893119 | 27893132 | phastConsElements100way | lod=108 | score:457, srow:3631854 |
| 34110 | chr17 | 27893134 | 27893138 | phastConsElements100way | lod=14 | score:255, srow:3631855 |
| 34110 | chr17 | 27893143 | 27893159 | phastConsElements100way | lod=76 | score:422, srow:3631856 |
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 34110 | cellmeth_recounts | PrEC: 90, LNCaP: 1780, DU145: 0 | 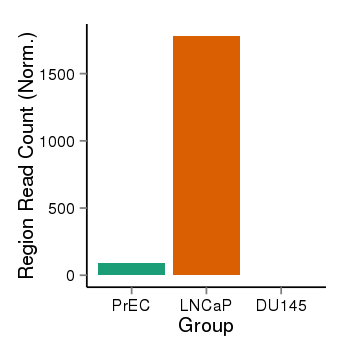 |
| 34110 | cellmeth_diffreps_PrECvsLNCaP | A=134, B=1782, Length=1400, Event=Up, log2FC=3.73, padj=0 | 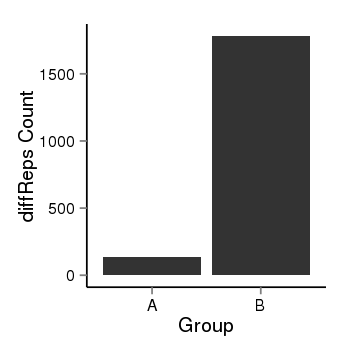 |
| 34110 | cellmeth_diffreps_PrECvsDU145 | A=133, B=0, Length=700, Event=Down, log2FC=-Inf, padj=0 | 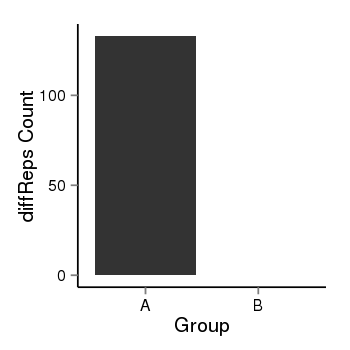 |
| 34110 | cellmeth_diffreps_LNCaPvsDU145 | A=1905.04, B=0.94, Length=1380, Event=Down, log2FC=-10.99, padj=0 | 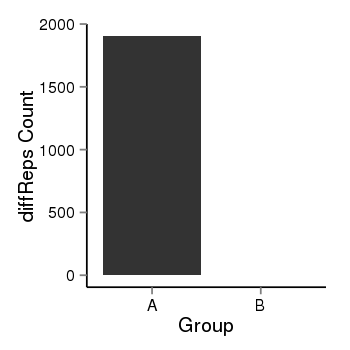 |
| 34110 | cellexp | id=ENSG00000167543, name=TP53I13, PrEC=1666, str=1928, LNCaP=2080, str=805, DU145=2005, str=1608 | 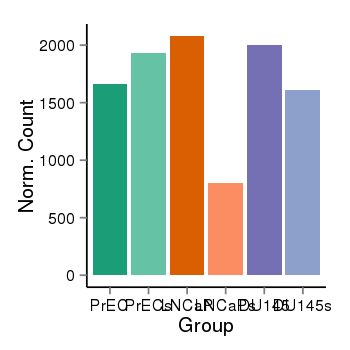 |
| 34110 | cellexp | id=ENSG00000168792, name=ABHD15, PrEC=672, str=1303, LNCaP=442, str=511, DU145=1346, str=1502 | 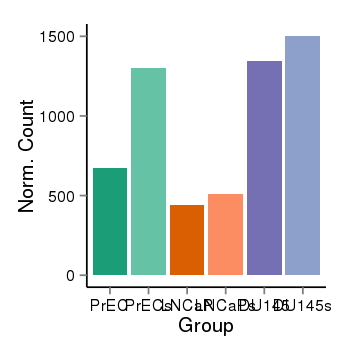 |
| 34110 | cellexp | id=ENSG00000264031, name=RP11-68I3.2, PrEC=0, str=0, LNCaP=0, str=0, DU145=0, str=0 |  |
| 34110 | tcgameth | site:cg04528771, B=0.45, L=0.62, H=0.59 | 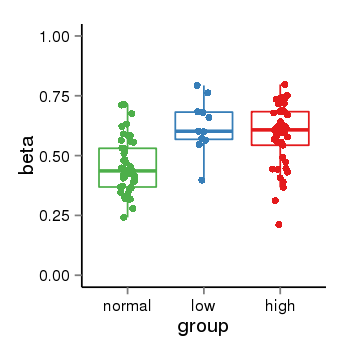 |
| 34110 | tcgameth | site:cg23112192, B=0.39, L=0.61, H=0.56 | 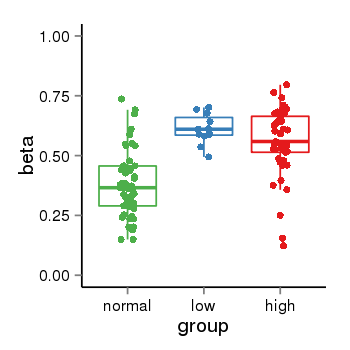 |
| 34110 | tcgameth | site:cg23473955, B=0.39, L=0.7, H=0.64 | 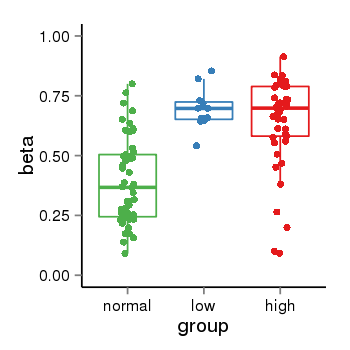 |
| 34110 | tcgaexp | gene=CRHR1, entrez=1394, pos=chr17:954315-43913194(+), B=36, L=21, M=25, H=18 | 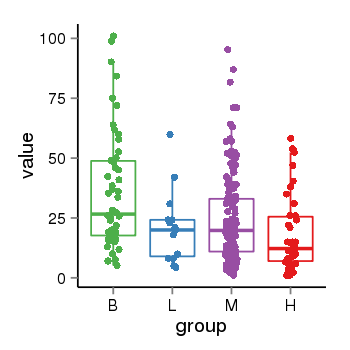 |
| 34110 | tcgaexp | gene=MAPT, entrez=4137, pos=chr17:762281-44105699(+), B=1933, L=2382, M=2487, H=2583 | 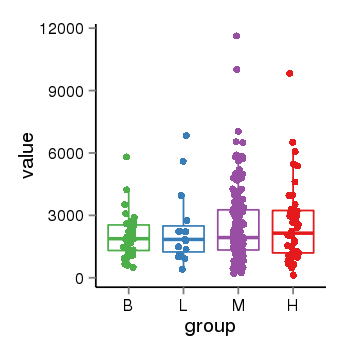 |
| 34110 | tcgaexp | gene=NSF, entrez=4905, pos=chr17:266212-44834828(+), B=3355, L=5678, M=5410, H=5700 | 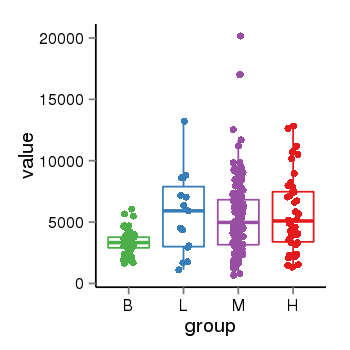 |
| 34110 | tcgaexp | gene=PLEKHM1, entrez=9842, pos=chr17:128328-43568146(-), B=1993, L=1631, M=1589, H=1418 | 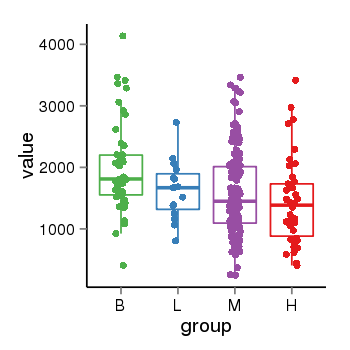 |
| 34110 | tcgaexp | gene=LRRC37A, entrez=9884, pos=chr17:415627-44415160(+), B=924, L=896, M=971, H=940 | 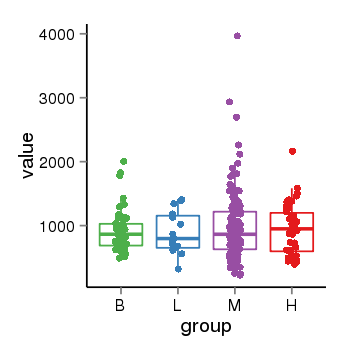 |
| 34110 | tcgaexp | gene=ARL17A, entrez=51326, pos=chr17:195721-44657088(-), B=200, L=195, M=210, H=230 | 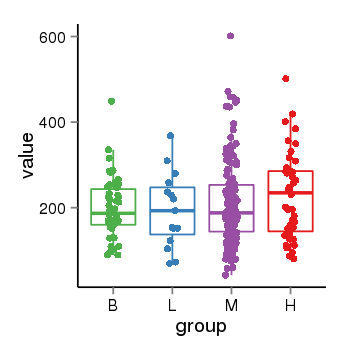 |
| 34110 | tcgaexp | gene=LRRC37A4P, entrez=55073, pos=chr17:197706-43597889(-), B=1015, L=1512, M=1396, H=1450 | 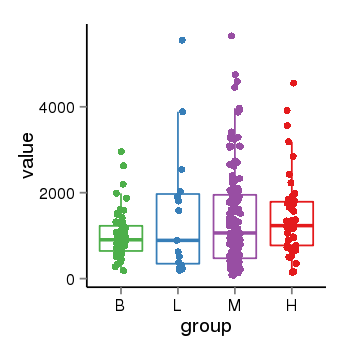 |
| 34110 | tcgaexp | gene=ABHD15, entrez=116236, pos=chr17:27887689-27894042(-), B=696, L=582, M=589, H=604 | 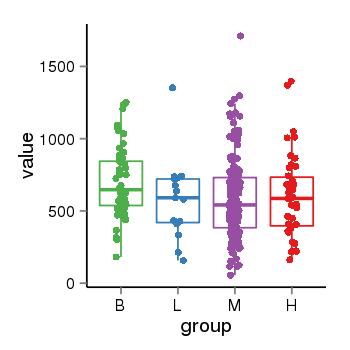 |
| 34110 | tcgaexp | gene=CRHR1-IT1, entrez=147081, pos=chr17:1143726-43723595(+), B=197, L=211, M=215, H=223 | 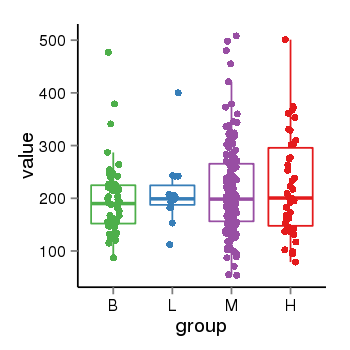 |
| 34110 | tcgaexp | gene=SPPL2C, entrez=162540, pos=chr17:943102-43924438(+), B=0, L=0, M=0, H=0 | 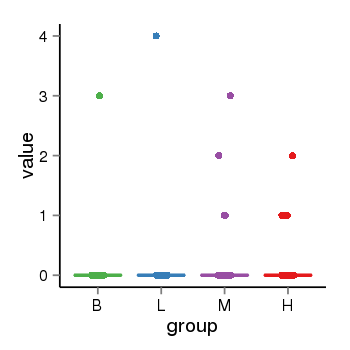 |
| 34110 | tcgaexp | gene=ARHGAP27, entrez=201176, pos=chr17:86404-43511112(-), B=1570, L=1264, M=1273, H=1486 | 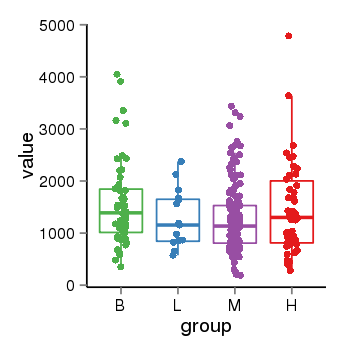 |
| 34110 | tcgaexp | gene=STH, entrez=246744, pos=chr17:790689-44077060(+), B=0, L=0, M=0, H=0 | 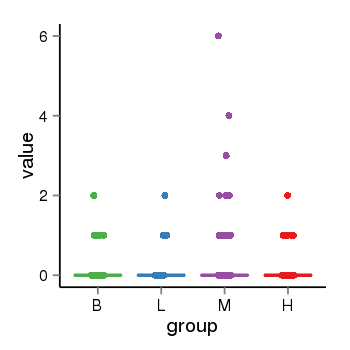 |
| 34110 | tcgaexp | gene=KANSL1, entrez=284058, pos=chr17:563498-44302740(-), B=3141, L=2594, M=2797, H=3017 | 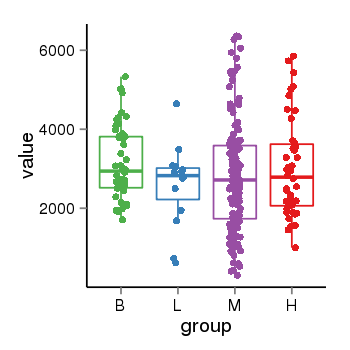 |
| 34110 | tcgaexp | gene=MGC57346, entrez=401884, pos=chr17:1151952-43715329(+), B=601, L=581, M=566, H=598 | 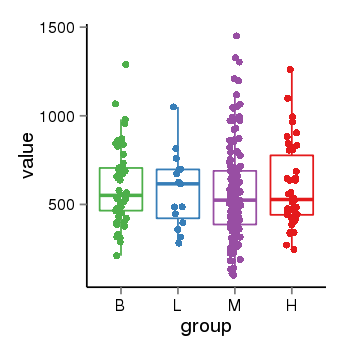 |
| 34110 | tcgaexp | gene=LRRC37A2, entrez=474170, pos=chr17:197706-44633014(+), B=351, L=310, M=361, H=377 | 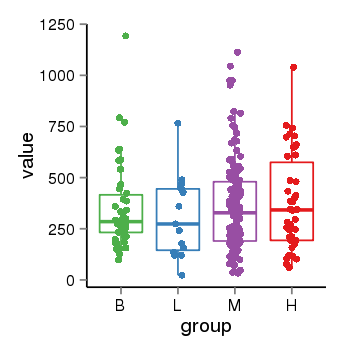 |
| 34110 | tcgaexp | gene=LOC644172, entrez=644172, pos=chr17:1188439-43679748(-), B=84, L=82, M=93, H=90 | 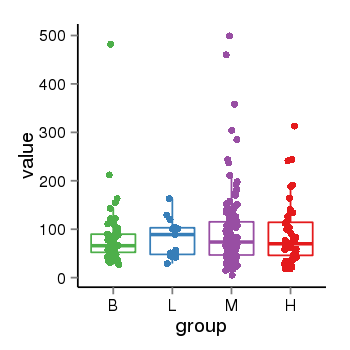 |
| 34110 | tcgaexp | gene=MAPT-AS1, entrez=100128977, pos=chr17:894694-43972879(-), B=1, L=2, M=4, H=2 | 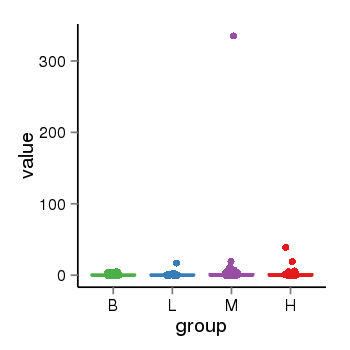 |
| 34110 | tcgaexp | gene=MAPT-IT1, entrez=100130148, pos=chr17:891434-43976164(+), B=5, L=3, M=3, H=4 | 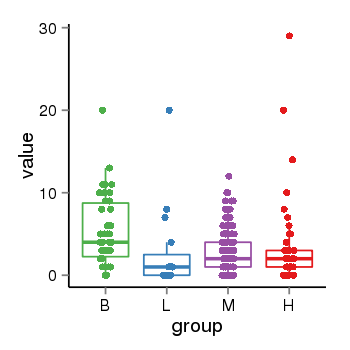 |