DMR 40255 chr12:75601451-75602000 (550bp)
| DMR Stats | |
|---|---|
| Position | chr12:75601451-75602000 (View on UCSC) |
| Width | 550bp |
| Genomic Context | exon |
| Nearest Genes | KCNC2 (0bp away) |
| ANODEV p-value | 6.16572055147e-07 |
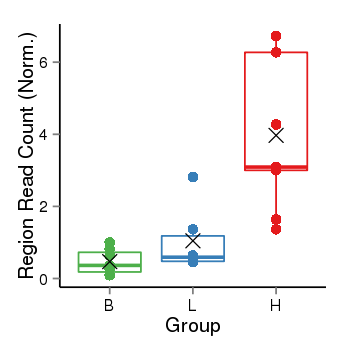
| Frequencies | Benign: 0.0 (0.0%), Low: 0.0 (0.0%), High: 6.0 (66.67 %) |
| Frequent? |
Region Count Boxplot
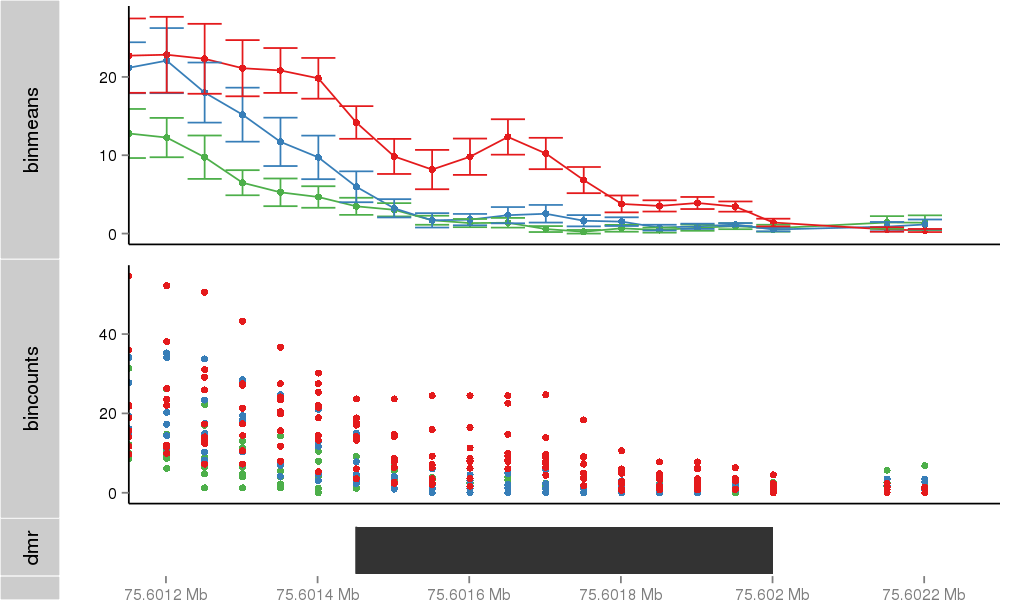
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 40255 | KCNC2 | 6759 | uc001sxe.4 | chr12 | 75433858 | 75603528 | - | 550 | 100.0 | 0.04 | 0 | 0.0 | E1 (0), I1 (39.82), E2 (60.36), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0), I5 (0), E6 (0) | 0.0 |
| 40255 | KCNC2 | 6759 | uc001sxf.4 | chr12 | 75433858 | 75603528 | - | 550 | 100.0 | 0.04 | 0 | 0.0 | E1 (0), I1 (39.82), E2 (60.36), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0) | 0.0 |
| 40255 | KCNC2 | 6759 | uc001sxg.2 | chr12 | 75433858 | 75603528 | - | 550 | 100.0 | 0.04 | 0 | 0.0 | E1 (0), I1 (39.82), E2 (60.36), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0) | 0.0 |
| 40255 | KCNC2 | 6759 | uc009zry.4 | chr12 | 75433858 | 75603528 | - | 550 | 100.0 | 0.08 | 0 | 0.0 | E1 (0), I1 (39.82), E2 (60.36), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0) | 0.0 |
| 40255 | KCNC2 | 6759 | uc010stw.2 | chr12 | 75433858 | 75603528 | - | 550 | 100.0 | 0.05 | 0 | 0.0 | E1 (0), I1 (39.82), E2 (60.36), I2 (0), E3 (0), I3 (0), E4 (0) | 0.0 |
| 40255 | KCNC2 | 6759 | uc031qif.1 | chr12 | 75433858 | 75603528 | - | 550 | 100.0 | 0.04 | 0 | 0.0 | E1 (0), I1 (39.82), E2 (60.36), I2 (0), E3 (0), I3 (0), E4 (0), I4 (0), E5 (0) | 0.0 |
- 6 geness
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 40255 | chr12 | 75601636 | 75601636 | cosmic | COSM312157 | srow:296545 |
| 40255 | chr12 | 75601636 | 75601636 | cosmic | COSM312156 | srow:296546 |
| 40255 | chr12 | 75601654 | 75601654 | cosmic | COSM115679 | srow:296547 |
| 40255 | chr12 | 75601666 | 75601666 | cosmic | COSM1211437 | srow:296548 |
| 40255 | chr12 | 75601666 | 75601666 | cosmic | COSM1211438 | srow:296549 |
| 40255 | chr12 | 75601678 | 75601678 | cosmic | COSM161845 | srow:296550 |
| 40255 | chr12 | 75601678 | 75601678 | cosmic | COSM161844 | srow:296551 |
| 40255 | chr12 | 75601680 | 75601681 | cosmic | COSM1513144 | srow:296552 |
| 40255 | chr12 | 75601680 | 75601681 | cosmic | COSM1513145 | srow:296553 |
| 40255 | chr12 | 75601696 | 75601696 | cosmic | COSM1364189 | srow:296554 |
| 40255 | chr12 | 75601696 | 75601696 | cosmic | COSM1364190 | srow:296555 |
| 40255 | chr12 | 75601697 | 75601697 | cosmic | COSM1364191 | srow:296556 |
| 40255 | chr12 | 75601697 | 75601697 | cosmic | COSM1364192 | srow:296557 |
| 40255 | chr12 | 75601712 | 75601712 | cosmic | COSM1179987 | srow:296558 |
| 40255 | chr12 | 75601712 | 75601712 | cosmic | COSM1179988 | srow:296559 |
| 40255 | chr12 | 75601717 | 75601717 | cosmic | COSM1513142 | srow:296560 |
| 40255 | chr12 | 75601717 | 75601717 | cosmic | COSM1513143 | srow:296561 |
| 40255 | chr12 | 75601734 | 75601734 | cosmic | COSM384264 | srow:296562 |
| 40255 | chr12 | 75601734 | 75601734 | cosmic | COSM384265 | srow:296563 |
| 40255 | chr12 | 75601441 | 75601463 | phastConsElements100way | lod=304 | score:559, srow:2085265 |
| 40255 | chr12 | 75601477 | 75601482 | phastConsElements100way | lod=18 | score:280, srow:2085266 |
| 40255 | chr12 | 75601507 | 75601516 | phastConsElements100way | lod=41 | score:361, srow:2085267 |
| 40255 | chr12 | 75601519 | 75601538 | phastConsElements100way | lod=112 | score:461, srow:2085268 |
| 40255 | chr12 | 75601549 | 75601554 | phastConsElements100way | lod=31 | score:333, srow:2085269 |
| 40255 | chr12 | 75601558 | 75601579 | phastConsElements100way | lod=43 | score:366, srow:2085270 |
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 40255 | cellmeth_recounts | PrEC: 1, LNCaP: 105, DU145: 22 | 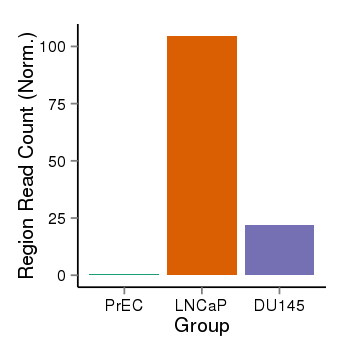 |
| 40255 | cellmeth_diffreps_PrECvsLNCaP | A=7, B=101, Length=680, Event=Up, log2FC=3.85, padj=0 | 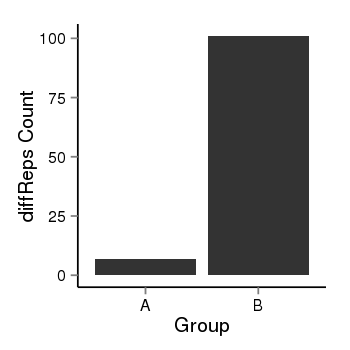 |
| 40255 | cellmeth_diffreps_LNCaPvsDU145 | A=110.11, B=17.77, Length=580, Event=Down, log2FC=-2.63, padj=0 | 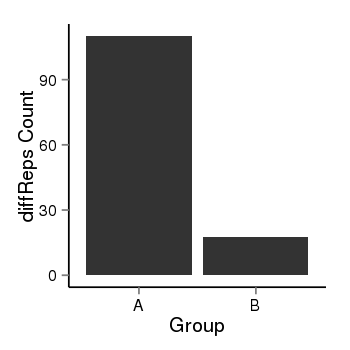 |
| 40255 | cellexp | id=ENSG00000166006, name=KCNC2, PrEC=0, str=0, LNCaP=11, str=12, DU145=0, str=0 | 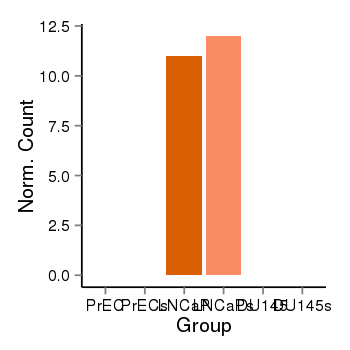 |
| 40255 | tcgameth | site:cg17203063, B=0.09, L=0.38, H=0.33 | 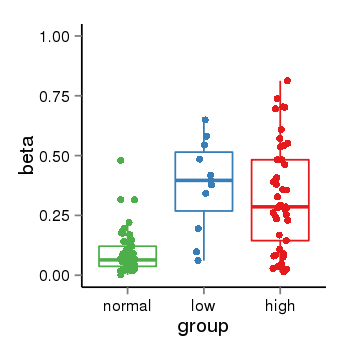 |
| 40255 | tcgameth | site:cg25820699, B=0.04, L=0.28, H=0.23 | 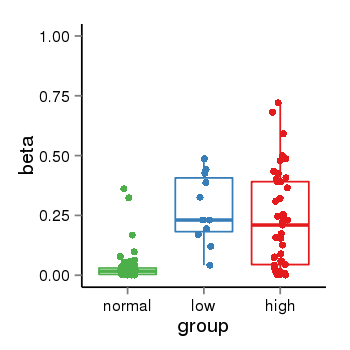 |
| 40255 | tcgaexp | gene=KCNC2, entrez=3747, pos=chr12:75433858-75603528(-), B=414, L=653, M=1571, H=3137 | 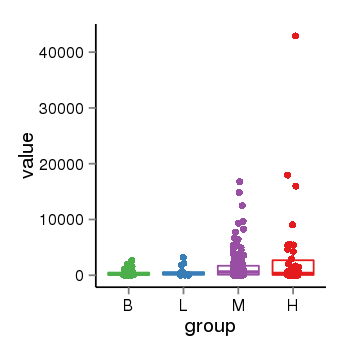 |
| 40255 | tcgaexp | gene=ZNF26, entrez=7574, pos=chr12:93510-133589154(+), B=419, L=392, M=412, H=512 | 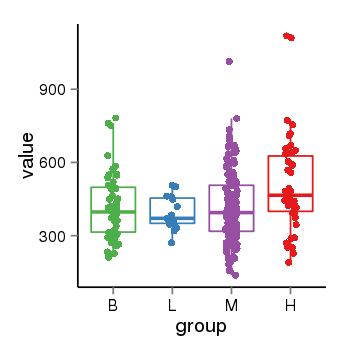 |
| 40255 | tcgaexp | gene=ZNF84, entrez=7637, pos=chr12:42780-133639885(+), B=1626, L=1649, M=1716, H=2045 | 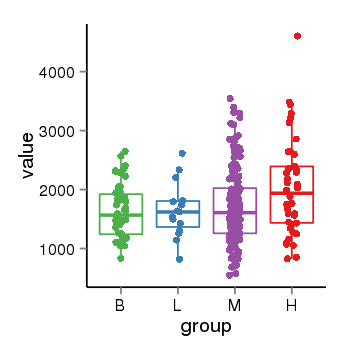 |
| 40255 | pubmed | gene=KCNC2, G=14, GP=0, GC=1, GM=0, GPM=0, GCM=0 | 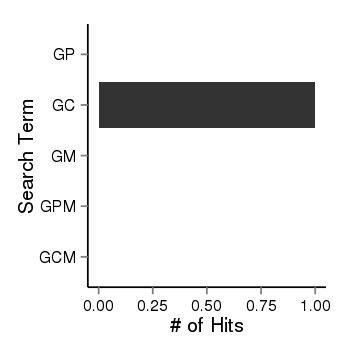 |