DMR 25951 chr2:45351-45700 (350bp)
| DMR Stats | |
|---|---|
| Position | chr2:45351-45700 (View on UCSC) |
| Width | 350bp |
| Genomic Context | exon |
| Nearest Genes | FAM110C (0bp away) |
| ANODEV p-value | 2.69284382914e-05 |
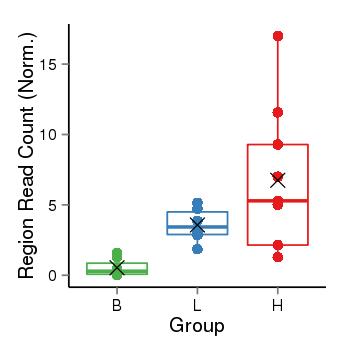
| Frequencies | Benign: 0.0 (0.0%), Low: 3.0 (50.0%), High: 6.0 (66.67 %) |
| Frequent? |
Region Count Boxplot
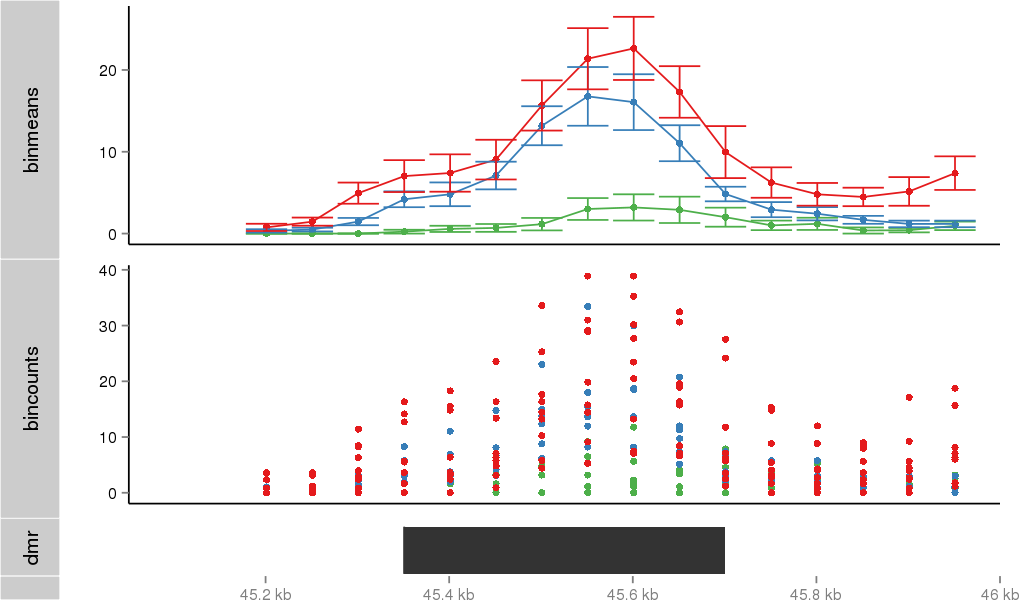
Region Overview
Gene Overlaps
| Dmrid | Gene Symbol | Gene Id | Isoform Id | Isoform Chr | Isoform Start | Isoform End | Isoform Strand | Overlap Bp | Query Overlap Per | Isoform Overlap Per | Noncoding | Promoter Per | Exonintron Per | End Per |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 25951 | FAM110C | 15961 | uc010yim.2 | chr2 | 38814 | 46588 | - | 350 | 100.0 | 0.01 | 0 | 0.0 | E1 (74.57), I1 (25.43), E2 (0) | 0.0 |
- 1 genes
Feature Overlaps
| Dmrid | Feature Chr | Feature Start | Feature End | Set | Name | Fields |
|---|---|---|---|---|---|---|
| 25951 | chr2 | 45532 | 45533 | phastConsElements100way | lod=38 | score:354, srow:4371899 |
| 25951 | chr2 | 45660 | 45662 | phastConsElements100way | lod=38 | score:354, srow:4371909 |
| 25951 | chr2 | 45550 | 45554 | phastConsElements100way | lod=41 | score:361, srow:4371902 |
| 25951 | chr2 | 45476 | 45483 | phastConsElements100way | lod=50 | score:381, srow:4371893 |
| 25951 | chr2 | 45604 | 45611 | phastConsElements100way | lod=50 | score:381, srow:4371906 |
| 25951 | chr2 | 45673 | 45683 | phastConsElements100way | lod=61 | score:400, srow:4371912 |
| 25951 | chr2 | 45565 | 45578 | phastConsElements100way | lod=74 | score:420, srow:4371904 |
- Previous
- Page 2 of 2
- 7 of 32 featuress
Omics Overlaps
| Dmrid | Omic Set | Data | Plot |
|---|---|---|---|
| 25951 | cellmeth_recounts | PrEC: 1, LNCaP: 107, DU145: 0 | 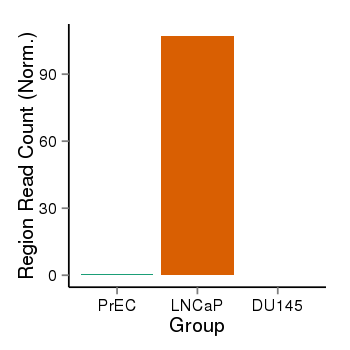 |
| 25951 | cellmeth_diffreps_PrECvsLNCaP | A=1, B=164, Length=1120, Event=Up, log2FC=7.36, padj=0 | 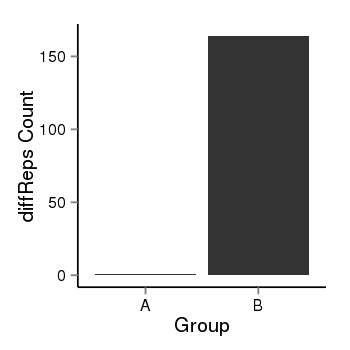 |
| 25951 | cellmeth_diffreps_LNCaPvsDU145 | A=175.32, B=0, Length=1120, Event=Down, log2FC=-Inf, padj=0 | 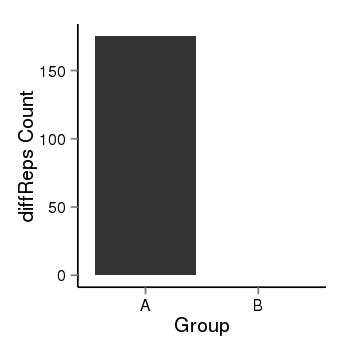 |
| 25951 | cellexp | id=ENSG00000184731, name=FAM110C, PrEC=4535, str=4854, LNCaP=224, str=138, DU145=780, str=633 | 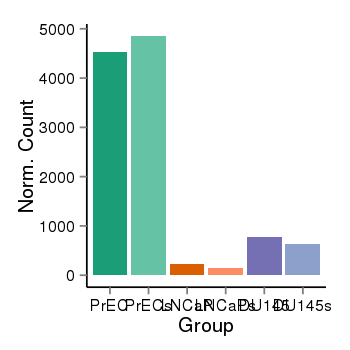 |
| 25951 | tcgameth | site:cg15237857, B=0.27, L=0.55, H=0.51 | 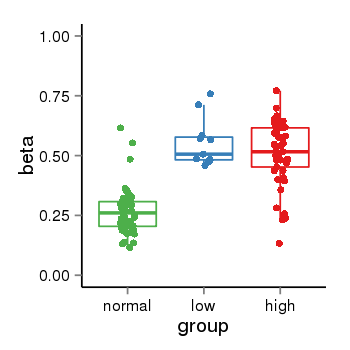 |
| 25951 | tcgaexp | gene=FAM110C, entrez=642273, pos=chr2:38814-46588(-), B=105, L=24, M=26, H=25 | 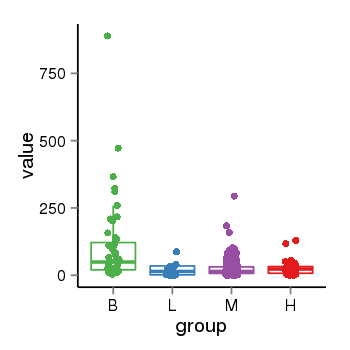 |